Bacterial chemoreceptor arrays are hexagonally packed trimers of receptor dimers networked by rings of kinase and coupling proteins
- PMID: 22355139
- PMCID: PMC3309718
- DOI: 10.1073/pnas.1115719109
Bacterial chemoreceptor arrays are hexagonally packed trimers of receptor dimers networked by rings of kinase and coupling proteins
Abstract
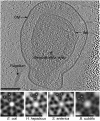
Chemoreceptor arrays are supramolecular transmembrane machines of unknown structure that allow bacteria to sense their surroundings and respond by chemotaxis. We have combined X-ray crystallography of purified proteins with electron cryotomography of native arrays inside cells to reveal the arrangement of the component transmembrane receptors, histidine kinases (CheA) and CheW coupling proteins. Trimers of receptor dimers lie at the vertices of a hexagonal lattice in a "two-facing-two" configuration surrounding a ring of alternating CheA regulatory domains (P5) and CheW couplers. Whereas the CheA kinase domains (P4) project downward below the ring, the CheA dimerization domains (P3) link neighboring rings to form an extended, stable array. This highly interconnected protein architecture underlies the remarkable sensitivity and cooperative nature of transmembrane signaling in bacterial chemotaxis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Mutational analysis of the chemoreceptor-coupling domain of the Escherichia coli chemotaxis signaling kinase CheA.J Bacteriol. 2006 May;188(9):3299-307. doi: 10.1128/JB.188.9.3299-3307.2006. J Bacteriol. 2006. PMID: 16621823 Free PMC article.
-
Core unit of chemotaxis signaling complexes.Proc Natl Acad Sci U S A. 2011 Jun 7;108(23):9390-5. doi: 10.1073/pnas.1104824108. Epub 2011 May 23. Proc Natl Acad Sci U S A. 2011. PMID: 21606342 Free PMC article.
-
New insights into bacterial chemoreceptor array structure and assembly from electron cryotomography.Biochemistry. 2014 Mar 18;53(10):1575-85. doi: 10.1021/bi5000614. Epub 2014 Mar 6. Biochemistry. 2014. PMID: 24580139 Free PMC article.
-
Stimulus response coupling in bacterial chemotaxis: receptor dimers in signalling arrays.Mol Microbiol. 1998 Nov;30(3):459-66. doi: 10.1046/j.1365-2958.1998.01066.x. Mol Microbiol. 1998. PMID: 9822812 Review.
-
Regulation of the chemotaxis histidine kinase CheA: A structural perspective.Biochim Biophys Acta Biomembr. 2020 Jan 1;1862(1):183030. doi: 10.1016/j.bbamem.2019.183030. Epub 2019 Jul 30. Biochim Biophys Acta Biomembr. 2020. PMID: 31374212 Free PMC article. Review.
Cited by
-
Attractant and repellent induce opposing changes in the four-helix bundle ligand-binding domain of a bacterial chemoreceptor.PLoS Biol. 2023 Dec 11;21(12):e3002429. doi: 10.1371/journal.pbio.3002429. eCollection 2023 Dec. PLoS Biol. 2023. PMID: 38079456 Free PMC article.
-
A divergent CheW confers plasticity to nucleoid-associated chemosensory arrays.PLoS Genet. 2019 Dec 20;15(12):e1008533. doi: 10.1371/journal.pgen.1008533. eCollection 2019 Dec. PLoS Genet. 2019. PMID: 31860666 Free PMC article.
-
Selective allosteric coupling in core chemotaxis signaling complexes.Proc Natl Acad Sci U S A. 2014 Nov 11;111(45):15940-5. doi: 10.1073/pnas.1415184111. Epub 2014 Oct 27. Proc Natl Acad Sci U S A. 2014. PMID: 25349385 Free PMC article.
-
Flexible Hinges in Bacterial Chemoreceptors.J Bacteriol. 2018 Feb 7;200(5):e00593-17. doi: 10.1128/JB.00593-17. Print 2018 Mar 1. J Bacteriol. 2018. PMID: 29229700 Free PMC article.
-
The nucleoid as a scaffold for the assembly of bacterial signaling complexes.PLoS Genet. 2017 Nov 21;13(11):e1007103. doi: 10.1371/journal.pgen.1007103. eCollection 2017 Nov. PLoS Genet. 2017. PMID: 29161263 Free PMC article.
References
-
- Kentner D, Sourjik V. Spatial organization of the bacterial chemotaxis system. Curr Opin Microbiol. 2006;9:619–624. - PubMed
-
- Wadhams GH, Armitage JP. Making sense of it all: Bacterial chemotaxis. Nat Rev Mol Cell Biol. 2004;5:1024–1037. - PubMed
-
- Sourjik V, Berg HC. Localization of components of the chemotaxis machinery of Escherichia coli using fluorescent protein fusions. Mol Microbiol. 2000;37:740–751. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

