Chromosome segregation in Archaea mediated by a hybrid DNA partition machine
- PMID: 22355141
- PMCID: PMC3309785
- DOI: 10.1073/pnas.1113384109
Chromosome segregation in Archaea mediated by a hybrid DNA partition machine
Abstract
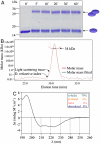
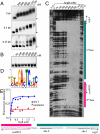
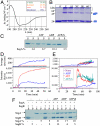
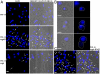
Eukarya and, more recently, some bacteria have been shown to rely on a cytoskeleton-based apparatus to drive chromosome segregation. In contrast, the factors and mechanisms underpinning this fundamental process are underexplored in archaea, the third domain of life. Here we establish that the archaeon Sulfolobus solfataricus harbors a hybrid segrosome consisting of two interacting proteins, SegA and SegB, that play a key role in genome segregation in this organism. SegA is an ortholog of bacterial, Walker-type ParA proteins, whereas SegB is an archaea-specific factor lacking sequence identity to either eukaryotic or bacterial proteins, but sharing homology with a cluster of uncharacterized factors conserved in both crenarchaea and euryarchaea, the two major archaeal sub-phyla. We show that SegA is an ATPase that polymerizes in vitro and that SegB is a site-specific DNA-binding protein contacting palindromic sequences located upstream of the segAB cassette. SegB interacts with SegA in the presence of nucleotides and dramatically affects its polymerization dynamics. Our data demonstrate that SegB strongly stimulates SegA polymerization, possibly by promoting SegA nucleation and accelerating polymer growth. Increased expression levels of segAB resulted in severe growth and chromosome segregation defects, including formation of anucleate cells, compact nucleoids confined to one half of the cell compartment and fragmented nucleoids. The overall picture emerging from our findings indicates that the SegAB complex fulfills a crucial function in chromosome segregation and is the prototype of a DNA partition machine widespread across archaea.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Kotwaliwale C, Biggins S. Microtubule capture: a concerted effort. Cell. 2006;127:1105–1108. - PubMed
-
- Hayes F, Barillà D. The bacterial segrosome: a dynamic nucleoprotein machine for DNA trafficking and segregation. Nat Rev Microbiol. 2006;4:133–143. - PubMed
-
- Schumacher MA. Structural biology of plasmid partition: uncovering the molecular mechanisms of DNA segregation. Biochem J. 2008;412:1–18. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

