The crucial role of PNPase in the degradation of small RNAs that are not associated with Hfq
- PMID: 22355164
- PMCID: PMC3312570
- DOI: 10.1261/rna.029413.111
The crucial role of PNPase in the degradation of small RNAs that are not associated with Hfq
Abstract
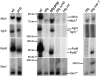
The transient existence of small RNAs free of binding to the RNA chaperone Hfq is part of the normal dynamic lifecycle of a sRNA. Small RNAs are extremely labile when not associated with Hfq, but the mechanism by which Hfq stabilizes sRNAs has been elusive. In this work we have found that polynucleotide phosphorylase (PNPase) is the major factor involved in the rapid degradation of small RNAs, especially those that are free of binding to Hfq. The levels of MicA, GlmY, RyhB, and SgrS RNAs are drastically increased upon PNPase inactivation in Hfq(-) cells. In the absence of Hfq, all sRNAs are slightly shorter than their full-length species as result of 3'-end trimming. We show that the turnover of Hfq-free small RNAs is growth-phase regulated, and that PNPase activity is particularly important in stationary phase. Indeed, PNPase makes a greater contribution than RNase E, which is commonly believed to be the main enzyme in the decay of small RNAs. Lack of poly(A) polymerase I (PAP I) is also found to affect the rapid degradation of Hfq-free small RNAs, although to a lesser extent. Our data also suggest that when the sRNA is not associated with Hfq, the degradation occurs mainly in a target-independent pathway in which RNase III has a reduced impact. This work demonstrated that small RNAs free of Hfq binding are preferably degraded by PNPase. Overall, our data highlight the impact of 3'-exonucleolytic RNA decay pathways and re-evaluates the degradation mechanisms of Hfq-free small RNAs.
Figures




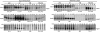
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
