Population genomics and phylogeography of an Australian dairy factory derived lytic bacteriophage
- PMID: 22355195
- PMCID: PMC3318435
- DOI: 10.1093/gbe/evs017
Population genomics and phylogeography of an Australian dairy factory derived lytic bacteriophage
Abstract
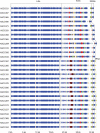
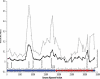
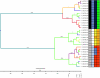
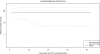
In this study, we present the full genomic sequences and evolutionary analyses of a serially sampled population of 28 Lactococcus lactis-infecting phage belonging to the 936-like group in Australia. Genome sizes were consistent with previously available genomes ranging in length from 30.9 to 32.1 Kbp and consisted of 55-65 open reading frames. We analyzed their genetic diversity and found that regions of high diversity are correlated with high recombination rate regions (P value = 0.01). Phylogenetic inference showed two major clades that correlate well with known host range. Using the extended Bayesian Skyline model, we found that population size has remained mostly constant through time. Moreover, the dispersion pattern of these genomes is in agreement with human-driven dispersion as suggested by phylogeographic analysis. In addition, selection analysis found evidence of positive selection on codon positions of the Receptor Binding Protein (RBP). Likewise, positively selected sites in the RBP were located within the neck and head region in the crystal structure, both known determinants of host range. Our study demonstrates the utility of phylogenetic methods applied to whole genome data collected from populations of phage for providing insights into applied microbiology.
Figures
References
-
- Akaike H. A new look at the statistical model identification. IEEE Trans Automat Contr. 1974;19:716–723.
-
- Boucher I, Emond E, Dion E, Montpetit D, Moineau S. Microbiological and molecular impacts of Abik on the lytic cycle of Lactococcus lactis phages of the 936 and P335 species. Microbiology. 2000;146:445–453. - PubMed
-
- Casjens SR. Comparative genomics and evolution of the tailed-bacteriophages. Curr Opin Microbiol. 2005;8:451–458. - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

