A comparison of rpoB and 16S rRNA as markers in pyrosequencing studies of bacterial diversity
- PMID: 22355318
- PMCID: PMC3280256
- DOI: 10.1371/journal.pone.0030600
A comparison of rpoB and 16S rRNA as markers in pyrosequencing studies of bacterial diversity
Abstract
Background: The 16S rRNA gene is the gold standard in molecular surveys of bacterial and archaeal diversity, but it has the disadvantages that it is often multiple-copy, has little resolution below the species level and cannot be readily interpreted in an evolutionary framework. We compared the 16S rRNA marker with the single-copy, protein-coding rpoB marker by amplifying and sequencing both from a single soil sample. Because the higher genetic resolution of the rpoB gene prohibits its use as a universal marker, we employed consensus-degenerate primers targeting the Proteobacteria.
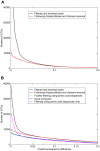
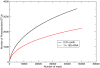
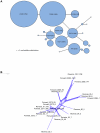
Methodology/principal findings: Pyrosequencing can be problematic because of the poor resolution of homopolymer runs. As these erroneous runs disrupt the reading frame of protein-coding sequences, removal of sequences containing nonsense mutations was found to be a valuable filter in addition to flowgram-based denoising. Although both markers gave similar estimates of total diversity, the rpoB marker revealed more species, requiring an order of magnitude fewer reads to obtain 90% of the true diversity. The application of population genetic methods was demonstrated on a particularly abundant sequence cluster.
Conclusions/significance: The rpoB marker can be a complement to the 16S rRNA marker for high throughput microbial diversity studies focusing on specific taxonomic groups. Additional error filtering is possible and tests for recombination or selection can be employed.
Conflict of interest statement
Figures
References
-
- Ward DM, Weller R, Bateson MM. 16S rRNA sequences reveal numerous uncultured microorganisms in a natural community. Nature. 1990;345:63–65. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

