A MAP6-related protein is present in protozoa and is involved in flagellum motility
- PMID: 22355359
- PMCID: PMC3280300
- DOI: 10.1371/journal.pone.0031344
A MAP6-related protein is present in protozoa and is involved in flagellum motility
Abstract
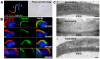
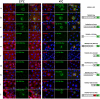
In vertebrates the microtubule-associated proteins MAP6 and MAP6d1 stabilize cold-resistant microtubules. Cilia and flagella have cold-stable microtubules but MAP6 proteins have not been identified in these organelles. Here, we describe TbSAXO as the first MAP6-related protein to be identified in a protozoan, Trypanosoma brucei. Using a heterologous expression system, we show that TbSAXO is a microtubule stabilizing protein. Furthermore we identify the domains of the protein responsible for microtubule binding and stabilizing and show that they share homologies with the microtubule-stabilizing Mn domains of the MAP6 proteins. We demonstrate, in the flagellated parasite, that TbSAXO is an axonemal protein that plays a role in flagellum motility. Lastly we provide evidence that TbSAXO belongs to a group of MAP6-related proteins (SAXO proteins) present only in ciliated or flagellated organisms ranging from protozoa to mammals. We discuss the potential roles of the SAXO proteins in cilia and flagella function.
Conflict of interest statement
Figures




References
-
- Amos LA, Schlieper D. Microtubules and maps. Adv Protein Chem. 2005;71:257–298. - PubMed
-
- Bosc C, Andrieux A, Job D. STOP proteins. Biochemistry. 2003;42:12125–12132. - PubMed
-
- Brinkley BR, Cartwright J., Jr Cold-labile and cold-stable microtubules in the mitotic spindle of mammalian cells. Ann N Y Acad Sci. 1975;253:428–439. - PubMed
-
- Gory-Faure S, Windscheid V, Bosc C, Peris L, Proietto D, et al. STOP-like protein 21 is a novel member of the STOP family, revealing a Golgi localization of STOP proteins. J Biol Chem. 2006;281:28387–28396. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

