Solexa sequencing identification of conserved and novel microRNAs in backfat of Large White and Chinese Meishan pigs
- PMID: 22355364
- PMCID: PMC3280305
- DOI: 10.1371/journal.pone.0031426
Solexa sequencing identification of conserved and novel microRNAs in backfat of Large White and Chinese Meishan pigs
Abstract
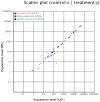
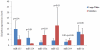
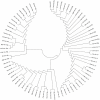
The domestic pig (Sus scrofa), an important species in animal production industry, is a right model for studying adipogenesis and fat deposition. In order to expand the repertoire of porcine miRNAs and further explore potential regulatory miRNAs which have influence on adipogenesis, high-throughput Solexa sequencing approach was adopted to identify miRNAs in backfat of Large White (lean type pig) and Meishan pigs (Chinese indigenous fatty pig). We identified 215 unique miRNAs comprising 75 known pre-miRNAs, of which 49 miRNA*s were first identified in our study, 73 miRNAs were overlapped in both libraries, and 140 were novelly predicted miRNAs, and 215 unique miRNAs were collectively corresponding to 235 independent genomic loci. Furthermore, we analyzed the sequence variations, seed edits and phylogenetic development of the miRNAs. 17 miRNAs were widely conserved from vertebrates to invertebrates, suggesting that these miRNAs may serve as potential evolutional biomarkers. 9 conserved miRNAs with significantly differential expressions were determined. The expression of miR-215, miR-135, miR-224 and miR-146b was higher in Large White pigs, opposite to the patterns shown by miR-1a, miR-133a, miR-122, miR-204 and miR-183. Almost all novel miRNAs could be considered pig-specific except ssc-miR-1343, miR-2320, miR-2326, miR-2411 and miR-2483 which had homologs in Bos taurus, among which ssc-miR-1343, miR-2320, miR-2411 and miR-2483 were validated in backfat tissue by stem-loop qPCR. Our results displayed a high level of concordance between the qPCR and Solexa sequencing method in 9 of 10 miRNAs comparisons except for miR-1a. Moreover, we found 2 miRNAs, miR-135 and miR-183, may exert impacts on porcine backfat development through WNT signaling pathway. In conclusion, our research develops porcine miRNAs and should be beneficial to study the adipogenesis and fat deposition of different pig breeds based on miRNAs.
Conflict of interest statement
Figures



References
-
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. - PubMed
-
- Pillai RS, Bhattacharyya SN, Filipowicz W. Repression of protein synthesis by miRNAs: how many mechanisms? Trends Cell Biol. 2007;17:118–126. - PubMed
-
- Zhang B, Pan X, Cobb GP, Anderson TA. Plant microRNA: a small regulatory molecule with big impact. Dev Biol. 2006;289:3–16. - PubMed
-
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. - PubMed
-
- Cullen BR. Viruses and microRNAs. Nat Genet. 2006;38:S25–S30. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

