Requirements for efficient proteolytic cleavage of prelamin A by ZMPSTE24
- PMID: 22355414
- PMCID: PMC3280227
- DOI: 10.1371/journal.pone.0032120
Requirements for efficient proteolytic cleavage of prelamin A by ZMPSTE24
Abstract
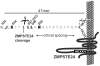
Background: The proteolytic maturation of the nuclear protein lamin A by the zinc metalloprotease ZMPSTE24 is critical for human health. The lamin A precursor, prelamin A, undergoes a multi-step maturation process that includes CAAX processing (farnesylation, proteolysis and carboxylmethylation of the C-terminal CAAX motif), followed by ZMPSTE24-mediated cleavage of the last 15 amino acids, including the modified C-terminus. Failure to cleave the prelamin A "tail", due to mutations in either prelamin A or ZMPSTE24, results in a permanently prenylated form of prelamin A that underlies the premature aging disease Hutchinson-Gilford Progeria Syndrome (HGPS) and related progeroid disorders.
Methodology/principal findings: Here we have investigated the features of the prelamin A substrate that are required for efficient cleavage by ZMPSTE24. We find that the C-terminal 41 amino acids of prelamin A contain sufficient context to allow cleavage of the tail by ZMPSTE24. We have identified several mutations in amino acids immediately surrounding the cleavage site (between Y646 and L647) that interfere with efficient cleavage of the prelamin A tail; these mutations include R644C, L648A and N650A, in addition to the previously reported L647R. Our data suggests that 9 of the 15 residues within the cleaved tail that lie immediately upstream of the CAAX motif are not critical for ZMPSTE24-mediated cleavage, as they can be replaced by the 9 amino acid HA epitope. However, duplication of the same 9 amino acids (to increase the distance between the prenyl group and the cleavage site) impairs the ability of ZMPSTE24 to cleave prelamin A.
Conclusions/significance: Our data reveals amino acid preferences flanking the ZMPSTE24 cleavage site of prelamin A and suggests that spacing from the farnesyl-cysteine to the cleavage site is important for optimal ZMPSTE24 cleavage. These studies begin to elucidate the substrate requirements of an enzyme activity critical to human health and longevity.
Conflict of interest statement
Figures
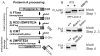

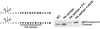

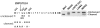
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

