Phylogenomic evidence for a common ancestor of mitochondria and the SAR11 clade
- PMID: 22355532
- PMCID: PMC3216501
- DOI: 10.1038/srep00013
Phylogenomic evidence for a common ancestor of mitochondria and the SAR11 clade
Abstract
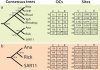
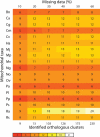
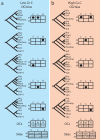
Mitochondria share a common ancestor with the Alphaproteobacteria, but determining their precise origins is challenging due to inherent difficulties in phylogenetically reconstructing ancient evolutionary events. Nonetheless, phylogenetic accuracy improves with more refined tools and expanded taxon sampling. We investigated mitochondrial origins with the benefit of new, deeply branching genome sequences from the ancient and prolific SAR11 clade of Alphaproteobacteria and publicly available alphaproteobacterial and mitochondrial genome sequences. Using the automated phylogenomic pipeline Hal, we systematically studied the effect of taxon sampling and missing data to accommodate small mitochondrial genomes. The evidence supports a common origin of mitochondria and SAR11 as a sister group to the Rickettsiales. The simplest explanation of these data is that mitochondria evolved from a planktonic marine alphaproteobacterial lineage that participated in multiple inter-specific cell colonization events, in some cases yielding parasitic relationships, but in at least one case producing a symbiosis that characterizes modern eukaryotic life.
Conflict of interest statement
All tree files, super alignments, and model files used in the study are available at: http://sar11-phylogenomics.cgrb.oregonstate.edu.
Figures




References
-
- Gray M. W., Lang B. F. & Burge G. Mitochondria of Protists. Annu. Rev. Genet. 38, 477–524 (2004). - PubMed
-
- Embley T. M. & Martin W. Eukaryotic evolution, changes and challenges. Nature 440, 623–630 (2006). - PubMed
-
- Fitzpatrick D. A., Creevey C. J. & McInerney J. O. Genome Phylogenies Indicate a Meaningful α-Proteobacterial Phylogeny and Support a Grouping of the Mitochondria with the Rickettsiales. Mol. Biol. Evol. 23, 74–85 (2005). - PubMed
-
- Andersson S. G. E. et al. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature 396, 133–143 (1998). - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

