A non-coding RNA of insect HzNV-1 virus establishes latent viral infection through microRNA
- PMID: 22355579
- PMCID: PMC3216547
- DOI: 10.1038/srep00060
A non-coding RNA of insect HzNV-1 virus establishes latent viral infection through microRNA
Abstract
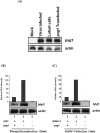
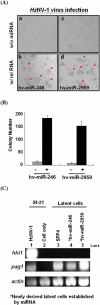
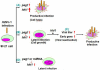
Heliothis zea nudivirus-1 (HzNV-1) is an insect virus previously known as Hz-1 baculovirus. One of its major early genes, hhi1, is responsible for the establishment of productive viral infection; another gene, pag1, which expresses a non-coding RNA, is the only viral transcript detectable during viral latency. Here we showed that this non-coding RNA was further processed into at least two distinct miRNAs, which targeted and degraded hhi1 transcript. This is a result strikingly similar to a recent report that herpes simplex virus produces tightly-regulated latent specific miRNAs to silence its own key early transcripts. Nevertheless, proof for the establishment of viral latency by miRNA is still lacking. We further showed that HzNV-1 latency could be directly induced by pag1-derived miRNAs in cells infected with a pag1-deleted, latency-deficient virus. This result suggests the existence of a novel mechanism, where miRNAs can be functional for the establishment of viral latency.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

