Tandem assembly of the epothilone biosynthetic gene cluster by in vitro site-specific recombination
- PMID: 22355658
- PMCID: PMC3216622
- DOI: 10.1038/srep00141
Tandem assembly of the epothilone biosynthetic gene cluster by in vitro site-specific recombination
Abstract
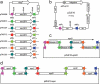
We describe a site-specific recombination-based tandem assembly (SSRTA) method for reconstruction of biological parts in synthetic biology. The system was catalyzed by Streptomyces phage φBT1 integrase, which belongs to the large serine recombinase subfamily. This one-step approach was efficient and accurate, and able to join multiple DNA molecules in vitro in a defined order. Thus, it could have applications in constructing metabolic pathways and genetic networks.
Figures
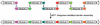

References
-
- Hillson N. J. in Design and Analysis of Biomolecular Circuits: Engineering Approaches to Systems and Synthetic Biology 295–314 (Springer-Verlag, Berlin, 2011).
-
- Ellis T., Adie T. & Baldwin G. S. DNA assembly for synthetic biology: from parts to pathways and beyond. Integr. Biol. 3, 109–118 (2011). - PubMed
-
- Smolke C. D. Building outside of the box: iGEM and the BioBricks Foundation. Nat. Biotechnol. 27, 1099–1102 (2009). - PubMed
-
- Elledge S. J. & Li M. Z. Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC. Nat. Methods 4, 251–256 (2007). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

