Mycobacterium tuberculosis Rv2419c, the missing glucosyl-3-phosphoglycerate phosphatase for the second step in methylglucose lipopolysaccharide biosynthesis
- PMID: 22355692
- PMCID: PMC3240985
- DOI: 10.1038/srep00177
Mycobacterium tuberculosis Rv2419c, the missing glucosyl-3-phosphoglycerate phosphatase for the second step in methylglucose lipopolysaccharide biosynthesis
Abstract
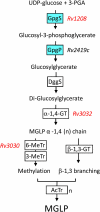
Mycobacteria synthesize intracellular methylglucose lipopolysaccharides (MGLP) proposed to regulate fatty acid synthesis. Although their structures have been elucidated, the identity of most biosynthetic genes remains unknown. The first step in MGLP biosynthesis is catalyzed by a glucosyl-3-phosphoglycerate synthase (GpgS, Rv1208 in Mycobacterium tuberculosis H37Rv). However, a typical glucosyl-3-phosphoglycerate phosphatase (GpgP, EC3.1.3.70) for dephosphorylation of glucosyl-3-phosphoglycerate to glucosylglycerate, was absent from mycobacterial genomes. We purified the native GpgP from Mycobacterium vanbaalenii and identified the corresponding gene deduced from amino acid sequences by mass spectrometry. The M. tuberculosis ortholog (Rv2419c), annotated as a putative phosphoglycerate mutase (PGM, EC5.4.2.1), was expressed and functionally characterized as a new GpgP. Regardless of the high specificity for glucosyl-3-phosphoglycerate, the mycobacterial GpgP is not a sequence homolog of known isofunctional GpgPs. The assignment of a new function in M. tuberculosis genome expands our understanding of this organism's genetic repertoire and of the early events in MGLP biosynthesis.
Figures

Similar articles
-
Identification of the mycobacterial glucosyl-3-phosphoglycerate synthase.FEMS Microbiol Lett. 2008 Mar;280(2):195-202. doi: 10.1111/j.1574-6968.2007.01064.x. Epub 2008 Jan 24. FEMS Microbiol Lett. 2008. PMID: 18221489
-
Mechanism of dephosphorylation of glucosyl-3-phosphoglycerate by a histidine phosphatase.J Biol Chem. 2014 Aug 1;289(31):21242-51. doi: 10.1074/jbc.M114.569913. Epub 2014 Jun 9. J Biol Chem. 2014. PMID: 24914210 Free PMC article.
-
Glucosylglycerate biosynthesis in the deepest lineage of the Bacteria: characterization of the thermophilic proteins GpgS and GpgP from Persephonella marina.J Bacteriol. 2007 Mar;189(5):1648-54. doi: 10.1128/JB.00841-06. Epub 2006 Dec 22. J Bacteriol. 2007. PMID: 17189358 Free PMC article.
-
Glucosylglycerate metabolism, bioversatility and mycobacterial survival.Glycobiology. 2017 Mar 4;27(3):213-227. doi: 10.1093/glycob/cww132. Glycobiology. 2017. PMID: 28025249 Review.
-
Genetics of Mycobacterial Arabinogalactan and Lipoarabinomannan Assembly.Microbiol Spectr. 2014 Aug;2(4):MGM2-0013-2013. doi: 10.1128/microbiolspec.MGM2-0013-2013. Microbiol Spectr. 2014. PMID: 26104198 Review.
Cited by
-
Structure of Mycobacterium thermoresistibile GlgE defines novel conformational states that contribute to the catalytic mechanism.Sci Rep. 2015 Nov 30;5:17144. doi: 10.1038/srep17144. Sci Rep. 2015. PMID: 26616850 Free PMC article.
-
Functional characterization of two members of histidine phosphatase superfamily in Mycobacterium tuberculosis.BMC Microbiol. 2013 Dec 11;13:292. doi: 10.1186/1471-2180-13-292. BMC Microbiol. 2013. PMID: 24330471 Free PMC article.
-
Mannosylglucosylglycerate biosynthesis in the deep-branching phylum Planctomycetes: characterization of the uncommon enzymes from Rhodopirellula baltica.Sci Rep. 2013;3:2378. doi: 10.1038/srep02378. Sci Rep. 2013. PMID: 23921581 Free PMC article.
-
Octanoylation of early intermediates of mycobacterial methylglucose lipopolysaccharides.Sci Rep. 2015 Sep 1;5:13610. doi: 10.1038/srep13610. Sci Rep. 2015. PMID: 26324178 Free PMC article.
-
Genome sequence of Mycobacterium hassiacum DSM 44199, a rare source of heat-stable mycobacterial proteins.J Bacteriol. 2012 Dec;194(24):7010-1. doi: 10.1128/JB.01880-12. J Bacteriol. 2012. PMID: 23209251 Free PMC article.
References
-
- Dye C. Doomsday postponed? Preventing and reversing epidemics of drug-resistant tuberculosis. Nat Rev Microbiol 7, 81–87 (2009). - PubMed
-
- Cole S. T., et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393, 537–544 (1998). - PubMed
-
- Smith W. L. & Ballou C. E. The 6-O-methylglucose-containing lipopolysaccharides of Mycobacterium phlei. Locations of the neutral and acidic acyl groups. J Biol Chem 248, 7118–7125 (1973). - PubMed
-
- Maitra S. K. & Ballou C. E. Heterogeneity and refined structures of 3-O-methyl-D-mannose polysaccharides from Mycobacterium smegmatis. J Biol Chem 252, 2459–2469 (1977). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

