Reconstruction and analysis of a genome-scale metabolic model for Scheffersomyces stipitis
- PMID: 22356827
- PMCID: PMC3310799
- DOI: 10.1186/1475-2859-11-27
Reconstruction and analysis of a genome-scale metabolic model for Scheffersomyces stipitis
Abstract
Background: Fermentation of xylose, the major component in hemicellulose, is essential for economic conversion of lignocellulosic biomass to fuels and chemicals. The yeast Scheffersomyces stipitis (formerly known as Pichia stipitis) has the highest known native capacity for xylose fermentation and possesses several genes for lignocellulose bioconversion in its genome. Understanding the metabolism of this yeast at a global scale, by reconstructing the genome scale metabolic model, is essential for manipulating its metabolic capabilities and for successful transfer of its capabilities to other industrial microbes.
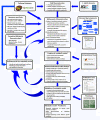
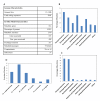
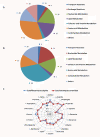
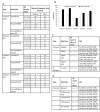
Results: We present a genome-scale metabolic model for Scheffersomyces stipitis, a native xylose utilizing yeast. The model was reconstructed based on genome sequence annotation, detailed experimental investigation and known yeast physiology. Macromolecular composition of Scheffersomyces stipitis biomass was estimated experimentally and its ability to grow on different carbon, nitrogen, sulphur and phosphorus sources was determined by phenotype microarrays. The compartmentalized model, developed based on an iterative procedure, accounted for 814 genes, 1371 reactions, and 971 metabolites. In silico computed growth rates were compared with high-throughput phenotyping data and the model could predict the qualitative outcomes in 74% of substrates investigated. Model simulations were used to identify the biosynthetic requirements for anaerobic growth of Scheffersomyces stipitis on glucose and the results were validated with published literature. The bottlenecks in Scheffersomyces stipitis metabolic network for xylose uptake and nucleotide cofactor recycling were identified by in silico flux variability analysis. The scope of the model in enhancing the mechanistic understanding of microbial metabolism is demonstrated by identifying a mechanism for mitochondrial respiration and oxidative phosphorylation.
Conclusion: The genome-scale metabolic model developed for Scheffersomyces stipitis successfully predicted substrate utilization and anaerobic growth requirements. Useful insights were drawn on xylose metabolism, cofactor recycling and mechanism of mitochondrial respiration from model simulations. These insights can be applied for efficient xylose utilization and cofactor recycling in other industrial microorganisms. The developed model forms a basis for rational analysis and design of Scheffersomyces stipitis metabolic network for the production of fuels and chemicals from lignocellulosic biomass.
Figures
Similar articles
-
Probing the bioethanol production potential of Scheffersomyces (Pichia) stipitis using validated genome-scale model.Biotechnol Lett. 2014 Dec;36(12):2443-51. doi: 10.1007/s10529-014-1629-8. Epub 2014 Aug 17. Biotechnol Lett. 2014. PMID: 25129048
-
Genome-scale metabolic reconstructions of Pichia stipitis and Pichia pastoris and in silico evaluation of their potentials.BMC Syst Biol. 2012 Apr 4;6:24. doi: 10.1186/1752-0509-6-24. BMC Syst Biol. 2012. PMID: 22472172 Free PMC article.
-
Elucidating redox balance shift in Scheffersomyces stipitis' fermentative metabolism using a modified genome-scale metabolic model.Microb Cell Fact. 2018 Sep 5;17(1):140. doi: 10.1186/s12934-018-0983-y. Microb Cell Fact. 2018. PMID: 30185188 Free PMC article.
-
Genetic improvement of native xylose-fermenting yeasts for ethanol production.J Ind Microbiol Biotechnol. 2015 Jan;42(1):1-20. doi: 10.1007/s10295-014-1535-z. Epub 2014 Nov 18. J Ind Microbiol Biotechnol. 2015. PMID: 25404205 Review.
-
Construction of advanced producers of first- and second-generation ethanol in Saccharomyces cerevisiae and selected species of non-conventional yeasts (Scheffersomyces stipitis, Ogataea polymorpha).J Ind Microbiol Biotechnol. 2020 Jan;47(1):109-132. doi: 10.1007/s10295-019-02242-x. Epub 2019 Oct 21. J Ind Microbiol Biotechnol. 2020. PMID: 31637550 Free PMC article. Review.
Cited by
-
Scheffersomyces stipitis: a comparative systems biology study with the Crabtree positive yeast Saccharomyces cerevisiae.Microb Cell Fact. 2012 Oct 9;11:136. doi: 10.1186/1475-2859-11-136. Microb Cell Fact. 2012. PMID: 23043429 Free PMC article.
-
Genome-scale modelling of microbial metabolism with temporal and spatial resolution.Biochem Soc Trans. 2015 Dec;43(6):1164-71. doi: 10.1042/BST20150146. Biochem Soc Trans. 2015. PMID: 26614655 Free PMC article. Review.
-
Dynamic metabolic modeling of a microaerobic yeast co-culture: predicting and optimizing ethanol production from glucose/xylose mixtures.Biotechnol Biofuels. 2013 Apr 1;6(1):44. doi: 10.1186/1754-6834-6-44. Biotechnol Biofuels. 2013. PMID: 23548183 Free PMC article.
-
Genome-scale modeling of yeast: chronology, applications and critical perspectives.FEMS Yeast Res. 2017 Aug 1;17(5):fox050. doi: 10.1093/femsyr/fox050. FEMS Yeast Res. 2017. PMID: 28899034 Free PMC article. Review.
-
Top-down and bottom-up microbiome engineering approaches to enable biomanufacturing from waste biomass.J Ind Microbiol Biotechnol. 2024 Jan 9;51:kuae025. doi: 10.1093/jimb/kuae025. J Ind Microbiol Biotechnol. 2024. PMID: 39003244 Free PMC article. Review.
References
-
- Kurtzman CP, Suzuki M. Phylogenetic analysis of the ascomycete yeasts that form coezyme Q-9 and the proposal of the new genera Babjeviella, Meyerozyma, Millerozyma, Priceomyces and Scheffersomyces. Mycoscience. 2010;51:2–14. doi: 10.1007/s10267-009-0011-5. - DOI
-
- du Preez JC, van Dreissel B, Prior BA. Ethanol tolerance of Pichia stipitis and Candida shehatae strains in fed-batch cultures at controlled low dissolved oxygen levels. Appl Microbiol Biotechnol. 1989;30:53–58.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

