Plant actin-binding protein SCAB1 is dimeric actin cross-linker with atypical pleckstrin homology domain
- PMID: 22356912
- PMCID: PMC3320945
- DOI: 10.1074/jbc.M111.338525
Plant actin-binding protein SCAB1 is dimeric actin cross-linker with atypical pleckstrin homology domain
Abstract
SCAB1 is a novel plant-specific actin-binding protein that binds, bundles, and stabilizes actin filaments and regulates stomatal movement. Here, we dissected the structure and function of SCAB1 by structural and biochemical approaches. We show that SCAB1 is composed of an actin-binding domain, two coiled-coil (CC) domains, and a fused immunoglobulin and pleckstrin homology (Ig-PH) domain. We determined crystal structures for the CC1 and Ig-PH domains at 1.9 and 1.7 Å resolution, respectively. The CC1 domain adopts an antiparallel helical hairpin that further dimerizes into a four-helix bundle. The CC2 domain also mediates dimerization. At least one of the coiled coils is required for actin binding, indicating that SCAB1 is a bivalent actin cross-linker. The key residues required for actin binding were identified. The PH domain lacks a canonical basic phosphoinositide-binding pocket but can bind weakly to inositol phosphates via a basic surface patch, implying the involvement of inositol signaling in SCAB1 regulation. Our results provide novel insights into the functional organization of SCAB1.
Figures





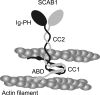
References
-
- Blanchoin L., Boujemaa-Paterski R., Henty J. L., Khurana P., Staiger C. J. (2010) Actin dynamics in plant cells: a team effort from multiple proteins orchestrates this very fast-paced game. Curr. Opin. Plant Biol. 13, 714–723 - PubMed
-
- Hussey P. J., Ketelaar T., Deeks M. J. (2006) Control of the actin cytoskeleton in plant cell growth. Annu. Rev. Plant Biol. 57, 109–125 - PubMed
-
- Thomas C., Tholl S., Moes D., Dieterle M., Papuga J., Moreau F., Steinmetz A. (2009) Actin bundling in plants. Cell Motil. Cytoskeleton 66, 940–957 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

