High-risk human papillomavirus E5 oncoprotein displays channel-forming activity sensitive to small-molecule inhibitors
- PMID: 22357280
- PMCID: PMC3347351
- DOI: 10.1128/JVI.06243-11
High-risk human papillomavirus E5 oncoprotein displays channel-forming activity sensitive to small-molecule inhibitors
Abstract
High-risk human papillomavirus type 16 (HPV16) is the primary causative agent of cervical cancer and therefore is responsible for significant morbidity and mortality worldwide. Cellular transformation is mediated directly by the expression of viral oncogenes, the least characterized of which, E5, subverts cellular proliferation and immune recognition processes. Despite a growing catalogue of E5-specific host interactions, little is understood regarding the molecular basis of its function. Here we describe a novel function for HPV16 E5 as an oligomeric channel-forming protein, placing it within the virus-encoded "viroporin" family. The development of a novel recombinant E5 expression system showed that E5 formed oligomeric assemblies of a defined luminal diameter and stoichiometry in membranous environments and that such channels mediated fluorescent dye release from liposomes. Hexameric E5 channel stoichiometry was suggested by native PAGE studies. In lieu of high-resolution structural information, established de novo molecular modeling and design methods permitted the development of the first specific small-molecule E5 inhibitor, capable of both abrogating channel activity in vitro and reducing E5-mediated effects on cell signaling pathways. The identification of channel activity should enhance the future understanding of the physiological function of E5 and could represent an important target for antiviral intervention.
Figures

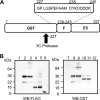
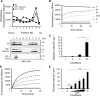



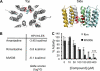

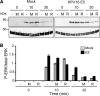
References
-
- Ashrafi GH, Haghshenas MR, Marchetti B, O'Brien PM, Campo MS. 2005. E5 protein of human papillomavirus type 16 selectively downregulates surface HLA class I. Int. J. Cancer 113:276–283 - PubMed
-
- Chang JL, et al. 2001. The expression of HPV-16 E5 protein in squamous neoplastic changes in the uterine cervix. J. Biomed. Sci. 8:206–213 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

