Breast cancer-associated Abraxas mutation disrupts nuclear localization and DNA damage response functions
- PMID: 22357538
- PMCID: PMC3869525
- DOI: 10.1126/scitranslmed.3003223
Breast cancer-associated Abraxas mutation disrupts nuclear localization and DNA damage response functions
Abstract
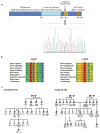
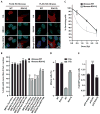
Breast cancer is the most common cancer in women in developed countries and has a well-established genetic component. Germline mutations in a network of genes encoding BRCA1, BRCA2, and their interacting partners confer hereditary susceptibility to breast cancer. Abraxas directly interacts with the BRCA1 BRCT (BRCA1 carboxyl-terminal) repeats and contributes to BRCA1-dependent DNA damage responses, making Abraxas a candidate for yet unexplained disease susceptibility. Here, we have screened 125 Northern Finnish breast cancer families for coding region and splice-site Abraxas mutations and genotyped three tagging single-nucleotide polymorphisms within the gene from 991 unselected breast cancer cases and 868 female controls for common cancer-associated variants. A novel heterozygous alteration, c.1082G>A (Arg361Gln), that results in abrogated nuclear localization and DNA response activities was identified in three breast cancer families and in one additional familial case from an unselected breast cancer cohort, but not in healthy controls (P = 0.002). On the basis of its exclusive occurrence in familial cancers, disease cosegregation, evolutionary conservation, and disruption of critical BRCA1 functions, the recurrent Abraxas c.1082G>A mutation connects to cancer predisposition. These findings contribute to the concept of a BRCA-centered tumor suppressor network and provide the identity of Abraxas as a new breast cancer susceptibility gene.
Conflict of interest statement
Present address: McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University School of Medicine, Broadway Research Building, 733 North Broadway, Baltimore, MD 21205, USA.
Figures

References
-
- Easton DF, Pooley KA, Dunning AM, Pharoah PD, Thompson D, Ballinger DG, Struewing JP, Morrison J, Field H, Luben R, Wareham N, Ahmed S, Healey CS, Bowma R, Meyer KB, Haiman CA, Kolonel LK, Henderson BE, Le Marchand L, Brennan P, Sangrajrang S, Gaborieau V, Odefrey F, Shen CY, Wu PE, Wang HC, Eccles D, Evans DG, Peto J, Fletcher O, Johnson N, Seal S, Stratton MR, Rahman N, Chenevix-Trench G, Bojesen SE, Nordestgaard BG, Axelsson CK, Garcia-Closas M, Bogdanova N, Schürmann P, Dörk T, Tollenaar RA, Jacobi CE, Devilee P, Klijn JG, Sigurdson AJ, Doody MM, Alexander BH, Zhang J, Cox A, Brock IW, MacPherson G, Reed MW, Couch FJ, Goode EL, Olson JE, Meijers-Heijboer H, van den Ouweland A, Uitterlinden A, Rivadeneira F, Milne RL, Ribas G, Gonzalez-Neira A, Benitez J, Hopper JL, McCredie M, Southey M, Giles GG, Schroen C, Justenhoven C, Brauch H, Hamann U, Ko YD, Spurdle AB, Beesley J, Chen X, Mannermaa A, Kosma VM, Kataja V, Hartikainen J, Day NE, Cox DR, Ponder BA SEARCH collaborators, kConFab AOCS Management Group. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–1093. - PMC - PubMed
-
- Stratton MR, Rahman N. The emerging landscape of breast cancer susceptibility. Nat Genet. 2008;40:17–22. - PubMed
-
- Walsh T, King MC. Ten genes for inherited breast cancer. Cancer Cell. 2007;11:103–105. - PubMed
-
- Shuen AY, Foulkes WD. Inherited mutations in breast cancer genes—Risk and response. J Mammary Gland Biol Neoplasia. 2011;16:3–15. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

