Full-length sequences of 11 hepatitis C virus genotype 2 isolates representing five subtypes and six unclassified lineages with unique geographical distributions and genetic variation patterns
- PMID: 22357752
- PMCID: PMC3755518
- DOI: 10.1099/vir.0.038315-0
Full-length sequences of 11 hepatitis C virus genotype 2 isolates representing five subtypes and six unclassified lineages with unique geographical distributions and genetic variation patterns
Abstract
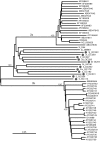
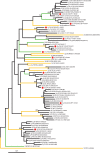
In this study, we characterized full-length hepatitis C virus (HCV) genome sequences for 11 genotype 2 isolates. They were isolated from the sera of 11 patients residing in Canada, of whom four had an African origin. Full-length genomes, each with 18-25 overlapping fragments, were obtained by PCR amplification. Five isolates represent the first complete genomes of subtypes 2d, 2e, 2j, 2m and 2r, while the other six correspond to variants that do not group within any assigned subtypes. These sequences had lengths of 9508-9825 nt and each contained a single ORF encoding 3012-3106 aa. Predicted amino acids were carefully inspected and unique variation patterns were recognized, especially for a 2e isolate, QC64. Phylogenetic analysis of complete genome sequences provides evidence that there are a total of 16 subtypes, of which 11 have been described here. Co-analysis with 68 partial NS5B sequences also differentiated 18 assigned subtypes, 2a-2r, and eight additional lineages within genotype 2, which is consistent with the analysis of complete genome sequences. The data from this study will now allow 10 assigned subtypes and six additional lineages of HCV genotype 2 to have their full-length genomes defined. Further analysis with 2021 genotype 2 sequences available in the HCV database indicated that the geographical distribution of these subtypes is consistent with an African origin, with particular subtypes having spread to Asia and the Americas.
Figures


References
-
- Abid K., Quadri R., Veuthey A. L., Hadengue A., Negro F. (2000). A novel hepatitis C virus (HCV) subtype from Somalia and its classification into HCV clade 3. J Gen Virol 81, 1485–1493 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

