A mutational study of Cnu reveals attractive forces between Cnu and H-NS
- PMID: 22358512
- PMCID: PMC3887714
- DOI: 10.1007/s10059-012-0006-5
A mutational study of Cnu reveals attractive forces between Cnu and H-NS
Abstract
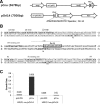
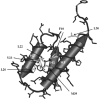
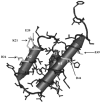
Cnu is a small 71-amino acid protein that complexes with H-NS and binds to a specific sequence in the replication origin of the E. coli chromosome. To understand the mechanism of interaction between Cnu and H-NS, we used bacterial genetics to select and analyze Cnu variants that cannot complex with H-NS. Out of 2,000 colonies, 40 Cnu variants were identified. Most variants (82.5%) had a single mutation, but a few variants (17.5%) had double amino acid changes. An in vitro assay was used to identify Cnu variants that were truly defective in H-NS binding. The changes in these defective variants occurred exclusively at charged amino acids (Asp, Glu, or Lys) on the surface of the protein. We propose that the attractive force that governs the Cnu-H-NS interaction is an ionic bond, unlike the hydrophobic interaction that is the major attractive force in most proteins.
Figures


References
-
- Badaut C., Williams R., Arluison V., Bouffartigues E., Robert B., Buc H., Rimsky S. The degree of oligomerization of the H-NS nucleoid structuring protein is related to specific binding to DNA. J. Biol. Chem. 2002;277:41657–41666. - PubMed
-
- Bae S.H., Liu D., Lim H.M., Lee Y., Choi B.S. Structure of the nucleoid-associated protein Cnu reveals common binding sites for H-NS in Cnu and Hha. Biochemistry. 2008;47:1993–2001. - PubMed
-
- Bloch V., Yang Y., Margeat E., Chavanieu A., Auge M.T., Robert B., Arold S., Rimsky S., Kochoyan M. The H-NS dimerization domain defines a new fold contributing to DNA recognition. Nat. Struct. Biol. 2003;10:212–218. - PubMed
-
- Bouffartigues E., Buckle M., Badaut C., Travers A., Rimsky S. H-NS cooperative binding to high-affinity sites in a regulatory element results in transcriptional silencing. Nat. Struct. Mol. Biol. 2007;14:441–448. - PubMed
-
- Cordeiro T.N., Garcia J., Pons J.I., Aznar S., Juarez A., Pons M. A single residue mutation in Hha preserving structure and binding to H-NS results in loss of H-NS mediated gene repression properties. FEBS Lett. 2008;582:3139–3144. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

