Dynamic energy landscapes of riboswitches help interpret conformational rearrangements and function
- PMID: 22359488
- PMCID: PMC3280964
- DOI: 10.1371/journal.pcbi.1002368
Dynamic energy landscapes of riboswitches help interpret conformational rearrangements and function
Abstract
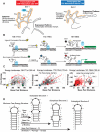
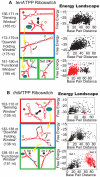
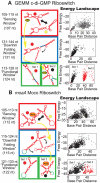
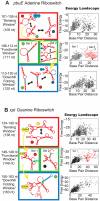
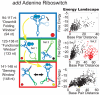
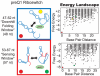
Riboswitches are RNAs that modulate gene expression by ligand-induced conformational changes. However, the way in which sequence dictates alternative folding pathways of gene regulation remains unclear. In this study, we compute energy landscapes, which describe the accessible secondary structures for a range of sequence lengths, to analyze the transcriptional process as a given sequence elongates to full length. In line with experimental evidence, we find that most riboswitch landscapes can be characterized by three broad classes as a function of sequence length in terms of the distribution and barrier type of the conformational clusters: low-barrier landscape with an ensemble of different conformations in equilibrium before encountering a substrate; barrier-free landscape in which a direct, dominant "downhill" pathway to the minimum free energy structure is apparent; and a barrier-dominated landscape with two isolated conformational states, each associated with a different biological function. Sharing concepts with the "new view" of protein folding energy landscapes, we term the three sequence ranges above as the sensing, downhill folding, and functional windows, respectively. We find that these energy landscape patterns are conserved in various riboswitch classes, though the order of the windows may vary. In fact, the order of the three windows suggests either kinetic or thermodynamic control of ligand binding. These findings help understand riboswitch structure/function relationships and open new avenues to riboswitch design.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

