The role of conserved waters in conformational transitions of Q61H K-ras
- PMID: 22359497
- PMCID: PMC3280954
- DOI: 10.1371/journal.pcbi.1002394
The role of conserved waters in conformational transitions of Q61H K-ras
Abstract
To investigate the stability and functional role of long-residence water molecules in the Q61H variant of the signaling protein K-ras, we analyzed all available Ras crystal structures and conformers derived from a series of independent explicit solvent molecular dynamics (MD) simulations totaling 1.76 µs. We show that the protein samples a different region of phase space in the presence and absence of several crystallographically conserved and buried water molecules. The dynamics of these waters is coupled with the local as well as the global motions of the protein, in contrast to less buried waters whose exchange with bulk is only loosely coupled with the motion of loops in their vicinity. Aided by two novel reaction coordinates involving the distance (d) between the C(α) atoms of G60 at switch 2 and G10 at the P-loop and the N-C(α)-C-O dihedral (ξ) of G60, we further show that three water molecules located in lobe1, at the interface between the lobes and at lobe2, are involved in the relative motion of residues at the two lobes of Q61H K-ras. Moreover, a d/ξ plot classifies the available Ras x-ray structures and MD-derived K-ras conformers into active GTP-, intermediate GTP-, inactive GDP-bound, and nucleotide-free conformational states. The population of these states and the transition between them is modulated by water-mediated correlated motions involving the functionally critical switch 2, P-loop and helix 3. These results suggest that water molecules act as allosteric ligands to induce a population shift among distinct switch 2 conformations that differ in effector recognition.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
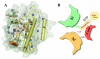
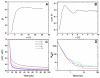
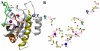
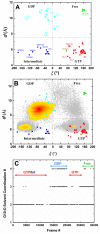
 (A);
(A);  (B);
(B);  (C);
(C);  (D);
(D);  (E); and
(E); and  (F). See legend of Fig. 4 for color scheme and Table 1 for simulation names.
(F). See legend of Fig. 4 for color scheme and Table 1 for simulation names.

Similar articles
-
DRoP: a water analysis program identifies Ras-GTP-specific pathway of communication between membrane-interacting regions and the active site.J Mol Biol. 2014 Feb 6;426(3):611-29. doi: 10.1016/j.jmb.2013.10.036. Epub 2013 Nov 2. J Mol Biol. 2014. PMID: 24189050
-
Nucleotide binding switches the information flow in ras GTPases.PLoS Comput Biol. 2011 Mar;7(3):e1001098. doi: 10.1371/journal.pcbi.1001098. Epub 2011 Mar 3. PLoS Comput Biol. 2011. PMID: 21390270 Free PMC article.
-
Analysis of binding site hot spots on the surface of Ras GTPase.J Mol Biol. 2011 Nov 4;413(4):773-89. doi: 10.1016/j.jmb.2011.09.011. Epub 2011 Sep 16. J Mol Biol. 2011. PMID: 21945529 Free PMC article.
-
Computational allosteric ligand binding site identification on Ras proteins.Acta Biochim Biophys Sin (Shanghai). 2016 Jan;48(1):3-10. doi: 10.1093/abbs/gmv100. Epub 2015 Oct 19. Acta Biochim Biophys Sin (Shanghai). 2016. PMID: 26487442 Review.
-
Ras Conformational Ensembles, Allostery, and Signaling.Chem Rev. 2016 Jun 8;116(11):6607-65. doi: 10.1021/acs.chemrev.5b00542. Epub 2016 Jan 27. Chem Rev. 2016. PMID: 26815308 Review.
Cited by
-
Mixed-Probe Simulation and Probe-Derived Surface Topography Map Analysis for Ligand Binding Site Identification.J Chem Theory Comput. 2017 Apr 11;13(4):1851-1861. doi: 10.1021/acs.jctc.7b00130. Epub 2017 Mar 13. J Chem Theory Comput. 2017. PMID: 28252958 Free PMC article.
-
Andrographolide derivatives inhibit guanine nucleotide exchange and abrogate oncogenic Ras function.Proc Natl Acad Sci U S A. 2013 Jun 18;110(25):10201-6. doi: 10.1073/pnas.1300016110. Epub 2013 Jun 4. Proc Natl Acad Sci U S A. 2013. PMID: 23737504 Free PMC article.
-
Oncogenic G12D mutation alters local conformations and dynamics of K-Ras.Sci Rep. 2019 Aug 13;9(1):11730. doi: 10.1038/s41598-019-48029-z. Sci Rep. 2019. PMID: 31409810 Free PMC article.
-
pMD-Membrane: A Method for Ligand Binding Site Identification in Membrane-Bound Proteins.PLoS Comput Biol. 2015 Oct 27;11(10):e1004469. doi: 10.1371/journal.pcbi.1004469. eCollection 2015 Oct. PLoS Comput Biol. 2015. PMID: 26506102 Free PMC article.
-
Comparison of the Conformations of KRAS Isoforms, K-Ras4A and K-Ras4B, Points to Similarities and Significant Differences.J Phys Chem B. 2016 Feb 4;120(4):667-79. doi: 10.1021/acs.jpcb.5b11110. Epub 2016 Jan 27. J Phys Chem B. 2016. PMID: 26761128 Free PMC article.
References
-
- Davey CA, Sargent DF, Luger K, Maeder AW, Richmond TJ. Solvent Mediated Interactions in the Structure of the Nucleosome Core Particle at 1.9 Å Resolution. J Mol Biol. 2002;319:1097–1113. - PubMed
-
- Mattos C. Protein water interactions in a dynamic world. Trends Biochem Sci. 2002;27:203–208. - PubMed
-
- Brunne RM, Liepinsh E, Otting G, Wüthrich K, van Gunsteren WF. Hydration of Proteins: A Comparison of Experimental Residence Times of Water Molecules Solvating the Bovine Pancreatic Trypsin Inhibitor with Theoretical Model Calculations. J Mol Biol. 1993;23:1040–1048. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

