Discovery of a modified tetrapolar sexual cycle in Cryptococcus amylolentus and the evolution of MAT in the Cryptococcus species complex
- PMID: 22359516
- PMCID: PMC3280970
- DOI: 10.1371/journal.pgen.1002528
Discovery of a modified tetrapolar sexual cycle in Cryptococcus amylolentus and the evolution of MAT in the Cryptococcus species complex
Abstract
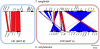
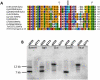
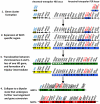
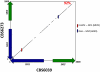
Sexual reproduction in fungi is governed by a specialized genomic region called the mating-type locus (MAT). The human fungal pathogenic and basidiomycetous yeast Cryptococcus neoformans has evolved a bipolar mating system (a, α) in which the MAT locus is unusually large (>100 kb) and encodes >20 genes including homeodomain (HD) and pheromone/receptor (P/R) genes. To understand how this unique bipolar mating system evolved, we investigated MAT in the closely related species Tsuchiyaea wingfieldii and Cryptococcus amylolentus and discovered two physically unlinked loci encoding the HD and P/R genes. Interestingly, the HD (B) locus sex-specific region is restricted (∼2 kb) and encodes two linked and divergently oriented homeodomain genes in contrast to the solo HD genes (SXI1α, SXI2a) of C. neoformans and Cryptococcus gattii. The P/R (A) locus contains the pheromone and pheromone receptor genes but has expanded considerably compared to other outgroup species (Cryptococcus heveanensis) and is linked to many of the genes also found in the MAT locus of the pathogenic Cryptococcus species. Our discovery of a heterothallic sexual cycle for C. amylolentus allowed us to establish the biological roles of the sex-determining regions. Matings between two strains of opposite mating-types (A1B1×A2B2) produced dikaryotic hyphae with fused clamp connections, basidia, and basidiospores. Genotyping progeny using markers linked and unlinked to MAT revealed that meiosis and uniparental mitochondrial inheritance occur during the sexual cycle of C. amylolentus. The sexual cycle is tetrapolar and produces fertile progeny of four mating-types (A1B1, A1B2, A2B1, and A2B2), but a high proportion of progeny are infertile, and fertility is biased towards one parental mating-type (A1B1). Our studies reveal insights into the plasticity and transitions in both mechanisms of sex determination (bipolar versus tetrapolar) and sexual reproduction (outcrossing versus inbreeding) with implications for similar evolutionary transitions and processes in fungi, plants, and animals.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Michod RE, Bernstein H, Nedelcu AM. Adaptive value of sex in microbial pathogens. Infection, Genetics and Evolution. 2008;8:267–285. - PubMed
-
- Heitman J. Sexual reproduction and the evolution of microbial pathogens. Current Biology. 2006;16:R711–725. - PubMed
-
- Fraser JA, Hsueh Y-P, Findley K, Heitman J. Evolution of the mating-type locus - the basidiomycetes. In: Heitman JWK J, Taylor JW, Casselton LA, editors. Sex in Fungi: molecular determination and evolutionary implications. Washington, DC: ASM Press; 2007. pp. 19–34.
-
- Lin X, Hull CM, Heitman J. Sexual reproduction between partners of the same mating type in Cryptococcus neoformans. Nature. 2005;434:1017–1021. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

