The origin and nature of tightly clustered BTG1 deletions in precursor B-cell acute lymphoblastic leukemia support a model of multiclonal evolution
- PMID: 22359517
- PMCID: PMC3280973
- DOI: 10.1371/journal.pgen.1002533
The origin and nature of tightly clustered BTG1 deletions in precursor B-cell acute lymphoblastic leukemia support a model of multiclonal evolution
Abstract
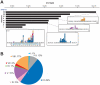
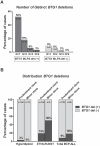
Recurrent submicroscopic deletions in genes affecting key cellular pathways are a hallmark of pediatric acute lymphoblastic leukemia (ALL). To gain more insight into the mechanism underlying these deletions, we have studied the occurrence and nature of abnormalities in one of these genes, the B-cell translocation gene 1 (BTG1), in a large cohort of pediatric ALL cases. BTG1 was found to be exclusively affected by genomic deletions, which were detected in 65 out of 722 B-cell precursor ALL (BCP-ALL) patient samples (9%), but not in 109 T-ALL cases. Eight different deletion sizes were identified, which all clustered at the telomeric site in a hotspot region within the second (and last) exon of the BTG1 gene, resulting in the expression of truncated BTG1 read-through transcripts. The presence of V(D)J recombination signal sequences at both sites of virtually all deletions strongly suggests illegitimate RAG1/RAG2-mediated recombination as the responsible mechanism. Moreover, high levels of histone H3 lysine 4 trimethylation (H3K4me3), which is known to tether the RAG enzyme complex to DNA, were found within the BTG1 gene body in BCP-ALL cells, but not T-ALL cells. BTG1 deletions were rarely found in hyperdiploid BCP-ALLs, but were predominant in other cytogenetic subgroups, including the ETV6-RUNX1 and BCR-ABL1 positive BCP-ALL subgroups. Through sensitive PCR-based screening, we identified multiple additional BTG1 deletions at the subclonal level in BCP-ALL, with equal cytogenetic distribution which, in some cases, grew out into the major clone at relapse. Taken together, our results indicate that BTG1 deletions may act as "drivers" of leukemogenesis in specific BCP-ALL subgroups, in which they can arise independently in multiple subclones at sites that are prone to aberrant RAG1/RAG2-mediated recombination events. These findings provide further evidence for a complex and multiclonal evolution of ALL.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Pui CH, Relling MV, Downing JR. Acute lymphoblastic leukemia. N Engl J Med. 2004;350:1535–1548. - PubMed
-
- Mullighan CG, Goorha S, Radtke I, Miller CB, Coustan-Smith E, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446:758–764. - PubMed
-
- Kuiper RP, Schoenmakers EF, Reijmersdal van SV, Hehir-Kwa JY, Geurts van Kessel A, et al. High-resolution genomic profiling of childhood ALL reveals novel recurrent genetic lesions affecting pathways involved in lymphocyte differentiation and cell cycle progression. Leukemia. 2007;21:1258–1266. - PubMed
-
- Kuiper RP, Waanders E, van der Velden VHJ, van Reijmersdal SV, Venkatachalam R, et al. IKZF1 deletions predict relapse in uniformly treated pediatric precursor B-ALL. Leukemia. 2010;24:1258–1264. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

