The retrohoming of linear group II intron RNAs in Drosophila melanogaster occurs by both DNA ligase 4-dependent and -independent mechanisms
- PMID: 22359518
- PMCID: PMC3280974
- DOI: 10.1371/journal.pgen.1002534
The retrohoming of linear group II intron RNAs in Drosophila melanogaster occurs by both DNA ligase 4-dependent and -independent mechanisms
Abstract
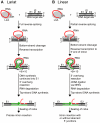
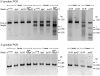
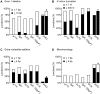
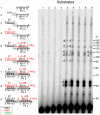
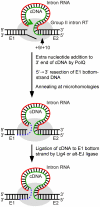
Mobile group II introns are bacterial retrotransposons that are thought to have invaded early eukaryotes and evolved into introns and retroelements in higher organisms. In bacteria, group II introns typically retrohome via full reverse splicing of an excised intron lariat RNA into a DNA site, where it is reverse transcribed by the intron-encoded protein. Recently, we showed that linear group II intron RNAs, which can result from hydrolytic splicing or debranching of lariat RNAs, can retrohome in eukaryotes by performing only the first step of reverse splicing, ligating their 3' end to the downstream DNA exon. Reverse transcription then yields an intron cDNA, whose free end is linked to the upstream DNA exon by an error-prone process that yields junctions similar to those formed by non-homologous end joining (NHEJ). Here, by using Drosophila melanogaster NHEJ mutants, we show that linear intron RNA retrohoming occurs by major Lig4-dependent and minor Lig4-independent mechanisms, which appear to be related to classical and alternate NHEJ, respectively. The DNA repair polymerase θ plays a crucial role in both pathways. Surprisingly, however, mutations in Ku70, which functions in capping chromosome ends during NHEJ, have only moderate, possibly indirect effects, suggesting that both Lig4 and the alternate end-joining ligase act in some retrohoming events independently of Ku. Another potential Lig4-independent mechanism, reverse transcriptase template switching from the intron RNA to the upstream exon DNA, occurs in vitro, but gives junctions differing from the majority in vivo. Our results show that group II introns can utilize cellular NHEJ enzymes for retromobility in higher organisms, possibly exploiting mechanisms that contribute to retrotransposition and mitigate DNA damage by resident retrotransposons. Additionally, our results reveal novel activities of group II intron reverse transcriptases, with implications for retrohoming mechanisms and potential biotechnological applications.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
Retrohoming of a Mobile Group II Intron in Human Cells Suggests How Eukaryotes Limit Group II Intron Proliferation.PLoS Genet. 2015 Aug 4;11(8):e1005422. doi: 10.1371/journal.pgen.1005422. eCollection 2015 Aug. PLoS Genet. 2015. PMID: 26241656 Free PMC article.
-
Linear group II intron RNAs can retrohome in eukaryotes and may use nonhomologous end-joining for cDNA ligation.Proc Natl Acad Sci U S A. 2009 Oct 27;106(43):18189-94. doi: 10.1073/pnas.0910277106. Epub 2009 Oct 15. Proc Natl Acad Sci U S A. 2009. PMID: 19833873 Free PMC article.
-
Genetic and biochemical assays reveal a key role for replication restart proteins in group II intron retrohoming.PLoS Genet. 2013 Apr;9(4):e1003469. doi: 10.1371/journal.pgen.1003469. Epub 2013 Apr 25. PLoS Genet. 2013. PMID: 23637634 Free PMC article.
-
Group II Intron RNPs and Reverse Transcriptases: From Retroelements to Research Tools.Cold Spring Harb Perspect Biol. 2019 Apr 1;11(4):a032375. doi: 10.1101/cshperspect.a032375. Cold Spring Harb Perspect Biol. 2019. PMID: 30936187 Free PMC article. Review.
-
Group II introns: mobile ribozymes that invade DNA.Cold Spring Harb Perspect Biol. 2011 Aug 1;3(8):a003616. doi: 10.1101/cshperspect.a003616. Cold Spring Harb Perspect Biol. 2011. PMID: 20463000 Free PMC article. Review.
Cited by
-
An optimized TALEN application for mutagenesis and screening in Drosophila melanogaster.Cell Logist. 2015 Feb 27;5(1):e1023423. doi: 10.1080/21592799.2015.1023423. eCollection 2015 Jan-Mar. Cell Logist. 2015. PMID: 26196022 Free PMC article.
-
Retrohoming of a Mobile Group II Intron in Human Cells Suggests How Eukaryotes Limit Group II Intron Proliferation.PLoS Genet. 2015 Aug 4;11(8):e1005422. doi: 10.1371/journal.pgen.1005422. eCollection 2015 Aug. PLoS Genet. 2015. PMID: 26241656 Free PMC article.
-
Mobile Bacterial Group II Introns at the Crux of Eukaryotic Evolution.Microbiol Spectr. 2015 Feb;3(1):MDNA3-0050-2014. doi: 10.1128/microbiolspec.MDNA3-0050-2014. Microbiol Spectr. 2015. PMID: 26104554 Free PMC article. Review.
-
Monoplacophoran mitochondrial genomes: convergent gene arrangements and little phylogenetic signal.BMC Evol Biol. 2016 Dec 16;16(1):274. doi: 10.1186/s12862-016-0829-3. BMC Evol Biol. 2016. PMID: 27986078 Free PMC article.
-
Genome Protection by DNA Polymerase θ.Annu Rev Genet. 2022 Nov 30;56:207-228. doi: 10.1146/annurev-genet-072920-041046. Epub 2022 Aug 26. Annu Rev Genet. 2022. PMID: 36028228 Free PMC article. Review.
References
-
- Martin W, Koonin EV. Introns and the origin of nucleus-cytosol compartmentalization. Nature. 2006;440:41–45. - PubMed
-
- Rodríguez-Trelles F, Tarrío R, Ayala FJ. Origins and evolution of spliceosomal introns. Annu Rev Genet. 2006;40:47–76. - PubMed
-
- Peebles CL, Perlman PS, Mecklenburg KL, Petrillo ML, Tabor JH, et al. A self-splicing RNA excises an intron lariat. Cell. 1986;44:213–223. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

