Small fragment homologous replacement: evaluation of factors influencing modification efficiency in an eukaryotic assay system
- PMID: 22359552
- PMCID: PMC3281040
- DOI: 10.1371/journal.pone.0030851
Small fragment homologous replacement: evaluation of factors influencing modification efficiency in an eukaryotic assay system
Abstract
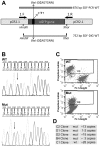
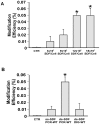
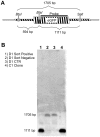
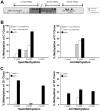
Homologous Replacement is used to modify specific gene sequences of chromosomal DNA in a process referred to as "Small Fragment Homologous Replacement", where DNA fragments replace genomic target resulting in specific sequence changes. To optimize the efficiency of this process, we developed a reporter based assay system where the replacement frequency is quantified by cytofluorimetric analysis following restoration of a stably integrated mutated eGFP gene in the genome of SV-40 immortalized mouse embryonic fibroblasts (MEF-SV-40). To obtain the highest correction frequency with this system, several parameters were considered: fragment synthesis and concentration, cell cycle phase and methylation status of both fragment and recipient genome. In addition, different drugs were employed to test their ability to improve technique efficiency. SFHR-mediated genomic modification resulted to be stably transmitted for several cell generations and confirmed at transcript and genomic levels. Modification efficiency was estimated in a range of 0.01-0.5%, further increasing when PARP-1 repair pathway was inhibited. In this study, for the first time SFHR efficiency issue was systematically approached and in part addressed, therefore opening new potential therapeutic ex-vivo applications.
Conflict of interest statement
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

