A novel protein kinase-like domain in a selenoprotein, widespread in the tree of life
- PMID: 22359664
- PMCID: PMC3281104
- DOI: 10.1371/journal.pone.0032138
A novel protein kinase-like domain in a selenoprotein, widespread in the tree of life
Abstract
Selenoproteins serve important functions in many organisms, usually providing essential oxidoreductase enzymatic activity, often for defense against toxic xenobiotic substances. Most eukaryotic genomes possess a small number of these proteins, usually not more than 20. Selenoproteins belong to various structural classes, often related to oxidoreductase function, yet a few of them are completely uncharacterised.Here, the structural and functional prediction for the uncharacterised selenoprotein O (SELO) is presented. Using bioinformatics tools, we predict that SELO protein adopts a three-dimensional fold similar to protein kinases. Furthermore, we argue that despite the lack of conservation of the "classic" catalytic aspartate residue of the archetypical His-Arg-Asp motif, SELO kinases might have retained catalytic phosphotransferase activity, albeit with an atypical active site. Lastly, the role of the selenocysteine residue is considered and the possibility of an oxidoreductase-regulated kinase function for SELO is discussed.The novel kinase prediction is discussed in the context of functional data on SELO orthologues in model organisms, FMP40 a.k.a.YPL222W (yeast), and ydiU (bacteria). Expression data from bacteria and yeast suggest a role in oxidative stress response. Analysis of genomic neighbourhoods of SELO homologues in the three domains of life points toward a role in regulation of ABC transport, in oxidative stress response, or in basic metabolism regulation. Among bacteria possessing SELO homologues, there is a significant over-representation of aquatic organisms, also of aerobic ones. The selenocysteine residue in SELO proteins occurs only in few members of this protein family, including proteins from Metazoa, and few small eukaryotes (Ostreococcus, stramenopiles). It is also demonstrated that enterobacterial mchC proteins involved in maturation of bactericidal antibiotics, microcins, form a distant subfamily of the SELO proteins.The new protein structural domain, with a putative kinase function assigned, expands the known kinome and deserves experimental determination of its biological role within the cell-signaling network.
Conflict of interest statement
Figures
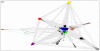
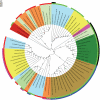



References
-
- Arner ES. Selenoproteins-What unique properties can arise with selenocysteine in place of cysteine? Exp Cell Res. 2010;316:1296–1303. - PubMed
-
- Kryukov GV, Castellano S, Novoselov SV, Lobanov AV, Zehtab O, et al. Characterization of mammalian selenoproteomes. Science. 2003;300:1439–1443. - PubMed
-
- Hanks SK, Quinn AM, Hunter T. The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science. 1988;241:42–52. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

