A whole-genome massively parallel sequencing analysis of BRCA1 mutant oestrogen receptor-negative and -positive breast cancers
- PMID: 22362584
- PMCID: PMC4976800
- DOI: 10.1002/path.4003
A whole-genome massively parallel sequencing analysis of BRCA1 mutant oestrogen receptor-negative and -positive breast cancers
Abstract
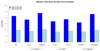
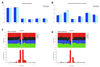
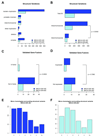
BRCA1 encodes a tumour suppressor protein that plays pivotal roles in homologous recombination (HR) DNA repair, cell-cycle checkpoints, and transcriptional regulation. BRCA1 germline mutations confer a high risk of early-onset breast and ovarian cancer. In more than 80% of cases, tumours arising in BRCA1 germline mutation carriers are oestrogen receptor (ER)-negative; however, up to 15% are ER-positive. It has been suggested that BRCA1 ER-positive breast cancers constitute sporadic cancers arising in the context of a BRCA1 germline mutation rather than being causally related to BRCA1 loss-of-function. Whole-genome massively parallel sequencing of ER-positive and ER-negative BRCA1 breast cancers, and their respective germline DNAs, was used to characterize the genetic landscape of BRCA1 cancers at base-pair resolution. Only BRCA1 germline mutations, somatic loss of the wild-type allele, and TP53 somatic mutations were recurrently found in the index cases. BRCA1 breast cancers displayed a mutational signature consistent with that caused by lack of HR DNA repair in both ER-positive and ER-negative cases. Sequencing analysis of independent cohorts of hereditary BRCA1 and sporadic non-BRCA1 breast cancers for the presence of recurrent pathogenic mutations and/or homozygous deletions found in the index cases revealed that DAPK3, TMEM135, KIAA1797, PDE4D, and GATA4 are potential additional drivers of breast cancers. This study demonstrates that BRCA1 pathogenic germline mutations coupled with somatic loss of the wild-type allele are not sufficient for hereditary breast cancers to display an ER-negative phenotype, and has led to the identification of three potential novel breast cancer genes (ie DAPK3, TMEM135, and GATA4).
Copyright © 2012 Pathological Society of Great Britain and Ireland. Published by John Wiley & Sons, Ltd.
Conflict of interest statement
Figures

Comment in
-
What drives breast cancer heterogeneity: oncogenic events or cell of origin?J Pathol. 2012 Jul;227(3):267-9. doi: 10.1002/path.4028. Epub 2012 May 8. J Pathol. 2012. PMID: 22431191
References
-
- Narod SA, Foulkes WD. BRCA1 and BRCA2: 1994 and beyond. Nat Rev Cancer. 2004;4:665–676. - PubMed
-
- Mullan PB, Quinn JE, Harkin DP. The role of BRCA1 in transcriptional regulation and cell cycle control. Oncogene. 2006;25:5854–5863. - PubMed
-
- Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. N Engl J Med. 2010;363:1938–1948. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

