Botulinum neurotoxin is shielded by NTNHA in an interlocked complex
- PMID: 22363010
- PMCID: PMC3545708
- DOI: 10.1126/science.1214270
Botulinum neurotoxin is shielded by NTNHA in an interlocked complex
Abstract
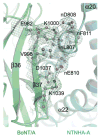
Botulinum neurotoxins (BoNTs) are highly poisonous substances that are also effective medicines. Accidental BoNT poisoning often occurs through ingestion of Clostridium botulinum-contaminated food. Here, we present the crystal structure of a BoNT in complex with a clostridial nontoxic nonhemagglutinin (NTNHA) protein at 2.7 angstroms. Biochemical and functional studies show that NTNHA provides large and multivalent binding interfaces to protect BoNT from gastrointestinal degradation. Moreover, the structure highlights key residues in BoNT that regulate complex assembly in a pH-dependent manner. Collectively, our findings define the molecular mechanisms by which NTNHA shields BoNT in the hostile gastrointestinal environment and releases it upon entry into the circulation. These results will assist in the design of small molecules for inhibiting oral BoNT intoxication and of delivery vehicles for oral administration of biologics.
Figures


Comment in
-
Structural biology. How a neurotoxin survives.Science. 2012 Feb 24;335(6071):928-9. doi: 10.1126/science.1219602. Science. 2012. PMID: 22362997 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

