Distribution, abundance, and diversity patterns of the thermoacidophilic "deep-sea hydrothermal vent euryarchaeota 2"
- PMID: 22363325
- PMCID: PMC3282477
- DOI: 10.3389/fmicb.2012.00047
Distribution, abundance, and diversity patterns of the thermoacidophilic "deep-sea hydrothermal vent euryarchaeota 2"
Abstract
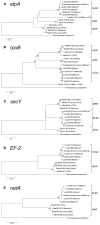
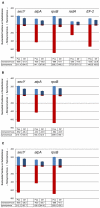
Cultivation-independent studies have shown that taxa belonging to the "deep-sea hydrothermal vent euryarchaeota 2" (DHVE2) lineage are widespread at deep-sea hydrothermal vents. While this lineage appears to be a common and important member of the microbial community at vent environments, relatively little is known about their overall distribution and phylogenetic diversity. In this study, we examined the distribution, relative abundance, co-occurrence patterns, and phylogenetic diversity of cultivable thermoacidophilic DHVE2 in deposits from globally distributed vent fields. Results of quantitative polymerase chain reaction assays with primers specific for the DHVE2 and Archaea demonstrate the ubiquity of the DHVE2 at deep-sea vents and suggest that they are significant members of the archaeal communities of established vent deposit communities. Local similarity analysis of pyrosequencing data revealed that the distribution of the DHVE2 was positively correlated with 10 other Euryarchaeota phylotypes and negatively correlated with mostly Crenarchaeota phylotypes. Targeted cultivation efforts resulted in the isolation of 12 axenic strains from six different vent fields, expanding the cultivable diversity of this lineage to vents along the East Pacific Rise and Mid-Atlantic Ridge. Eleven of these isolates shared greater than 97% 16S rRNA gene sequence similarity with one another and the only described isolate of the DHVE2, Aciduliprofundum boonei T469(T). Sequencing and phylogenetic analysis of five protein-coding loci, atpA, EF-2, radA, rpoB, and secY, revealed clustering of isolates according to geographic region of isolation. Overall, this study increases our understanding of the distribution, abundance, and phylogenetic diversity of the DHVE2.
Keywords: acidophile; archaea; biogeography; deep-sea; hydrothermal vents; multi-locus sequence analysis; thermophile.
Figures





References
-
- Bonch-Osmolovskaya E. A., Stetter K. O. (1991). Interspecies hydrogen transfer in cocultures of thermophilic archaea. Syst. Appl. Microbiol. 14, 205–20810.1016/S0723-2020(11)80369-3 - DOI
-
- Burggraf S., Jannasch H., Nicolaus B., Stetter K. (1990). Archaeoglobus profundus sp. nov., represents a new species within the sulfate-reducing archaebacteria. Syst. Appl. Microbiol. 13, 24–2810.1016/S0723-2020(11)80197-9 - DOI
LinkOut - more resources
Full Text Sources

