A comparative study of human TLR 7/8 stimulatory trimer compositions in influenza A viral genomes
- PMID: 22363482
- PMCID: PMC3281872
- DOI: 10.1371/journal.pone.0030751
A comparative study of human TLR 7/8 stimulatory trimer compositions in influenza A viral genomes
Abstract
Background: Variation in the genomes of single-stranded RNA viruses affects their infectivity and pathogenicity in two ways. First, viral genome sequence variations lead to changes in viral protein sequences and activities. Second, viral genome sequence variation produces diversity at the level of nucleotide composition and diversity in the interactions between viral RNAs and host toll-like receptors (TLRs). A viral genome-typing method based on this type of diversity has not yet been established.
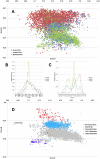
Methodology/principal findings: In this study, we propose a novel genomic trait called the "TLR stimulatory trimer composition" (TSTC) and two quantitative indicators, Score S and Score N, named "TLR stimulatory scores" (TSS). Using the complete genome sequences of 10,994 influenza A viruses (IAV) and 251 influenza B viruses, we show that TSTC analysis reveals the diversity of Score S and Score N among the IAVs isolated from various hosts. In addition, we show that low values of Score S are correlated with high pathogenicity and pandemic potential in IAVs. Finally, we use Score S and Score N to construct a logistic regression model to recognize IAV strains that are highly pathogenic or have high pandemic potential.
Conclusions/significance: Results from the TSTC analysis indicate that there are large differences between human and avian IAV genomes (except for segment 3), as illustrated by Score S. Moreover, segments 1, 2, 3 and 4 may be major determinants of the stimulatory activity exerted on human TLRs 7 and 8. We also find that a low Score S value is associated with high pathogenicity and pandemic potential in IAV. The π value from the TSS-derived logistic regression model is useful for recognizing emerging IAVs that have high pathogenicity and pandemic potential.
Conflict of interest statement
Figures






Similar articles
-
Efficient Inhibition of Avian and Seasonal Influenza A Viruses by a Virus-Specific Dicer-Substrate Small Interfering RNA Swarm in Human Monocyte-Derived Macrophages and Dendritic Cells.J Virol. 2019 Feb 5;93(4):e01916-18. doi: 10.1128/JVI.01916-18. Print 2019 Feb 15. J Virol. 2019. PMID: 30463970 Free PMC article.
-
Wastewater monitoring of human and avian influenza A viruses in Northern Ireland: a genomic surveillance study.Lancet Microbe. 2024 Dec;5(12):100933. doi: 10.1016/S2666-5247(24)00175-7. Epub 2024 Oct 9. Lancet Microbe. 2024. PMID: 39395428
-
Zoonotic Potential of Influenza A Viruses: A Comprehensive Overview.Viruses. 2018 Sep 13;10(9):497. doi: 10.3390/v10090497. Viruses. 2018. PMID: 30217093 Free PMC article. Review.
-
Genomic surveillance of avian-origin influenza A viruses causing human disease.Genome Med. 2018 Jun 27;10(1):50. doi: 10.1186/s13073-018-0560-3. Genome Med. 2018. PMID: 29950176 Free PMC article. Review.
-
Tissue Tropisms of Avian Influenza A Viruses Affect Their Spillovers from Wild Birds to Pigs.J Virol. 2020 Nov 23;94(24):e00847-20. doi: 10.1128/JVI.00847-20. Print 2020 Nov 23. J Virol. 2020. PMID: 32967956 Free PMC article.
Cited by
-
Viruses Seen by Our Cells: The Role of Viral RNA Sensors.J Immunol Res. 2018 Apr 30;2018:9480497. doi: 10.1155/2018/9480497. eCollection 2018. J Immunol Res. 2018. PMID: 29854853 Free PMC article. Review.
-
Favipiravir, an anti-influenza drug against life-threatening RNA virus infections.Pharmacol Ther. 2020 May;209:107512. doi: 10.1016/j.pharmthera.2020.107512. Epub 2020 Feb 22. Pharmacol Ther. 2020. PMID: 32097670 Free PMC article. Review.
-
Effects of Gui Zhi Ma Huang Ge Ban Tang on the TLR7 Pathway in Influenza Virus Infected Mouse Lungs in a Cold Environment.Evid Based Complement Alternat Med. 2018 Apr 23;2018:5939720. doi: 10.1155/2018/5939720. eCollection 2018. Evid Based Complement Alternat Med. 2018. PMID: 29849712 Free PMC article.
-
Low compositions of human toll-like receptor 7/8-stimulating RNA motifs in the MERS-CoV, SARS-CoV and SARS-CoV-2 genomes imply a substantial ability to evade human innate immunity.PeerJ. 2021 Feb 24;9:e11008. doi: 10.7717/peerj.11008. eCollection 2021. PeerJ. 2021. PMID: 33665043 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

