iNR-PhysChem: a sequence-based predictor for identifying nuclear receptors and their subfamilies via physical-chemical property matrix
- PMID: 22363503
- PMCID: PMC3283608
- DOI: 10.1371/journal.pone.0030869
iNR-PhysChem: a sequence-based predictor for identifying nuclear receptors and their subfamilies via physical-chemical property matrix
Abstract
Nuclear receptors (NRs) form a family of ligand-activated transcription factors that regulate a wide variety of biological processes, such as homeostasis, reproduction, development, and metabolism. Human genome contains 48 genes encoding NRs. These receptors have become one of the most important targets for therapeutic drug development. According to their different action mechanisms or functions, NRs have been classified into seven subfamilies. With the avalanche of protein sequences generated in the postgenomic age, we are facing the following challenging problems. Given an uncharacterized protein sequence, how can we identify whether it is a nuclear receptor? If it is, what subfamily it belongs to? To address these problems, we developed a predictor called iNR-PhysChem in which the protein samples were expressed by a novel mode of pseudo amino acid composition (PseAAC) whose components were derived from a physical-chemical matrix via a series of auto-covariance and cross-covariance transformations. It was observed that the overall success rate achieved by iNR-PhysChem was over 98% in identifying NRs or non-NRs, and over 92% in identifying NRs among the following seven subfamilies: NR1--thyroid hormone like, NR2--HNF4-like, NR3--estrogen like, NR4--nerve growth factor IB-like, NR5--fushi tarazu-F1 like, NR6--germ cell nuclear factor like, and NR0--knirps like. These rates were derived by the jackknife tests on a stringent benchmark dataset in which none of protein sequences included has ≥60% pairwise sequence identity to any other in a same subset. As a user-friendly web-server, iNR-PhysChem is freely accessible to the public at either http://www.jci-bioinfo.cn/iNR-PhysChem or http://icpr.jci.edu.cn/bioinfo/iNR-PhysChem. Also a step-by-step guide is provided on how to use the web-server to get the desired results without the need to follow the complicated mathematics involved in developing the predictor. It is anticipated that iNR-PhysChem may become a useful high throughput tool for both basic research and drug design.
Conflict of interest statement
Figures

 unit. (b) The cross-covariance refers to the coupling between two subsequences from two different sequences as indicated by two open curly braces.
unit. (b) The cross-covariance refers to the coupling between two subsequences from two different sequences as indicated by two open curly braces.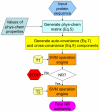

Similar articles
-
NR-2L: a two-level predictor for identifying nuclear receptor subfamilies based on sequence-derived features.PLoS One. 2011;6(8):e23505. doi: 10.1371/journal.pone.0023505. Epub 2011 Aug 15. PLoS One. 2011. PMID: 21858146 Free PMC article.
-
Recent progresses in identifying nuclear receptors and their families.Curr Top Med Chem. 2013;13(10):1192-200. doi: 10.2174/15680266113139990006. Curr Top Med Chem. 2013. PMID: 23647541 Review.
-
Application of Machine Learning Methods in Predicting Nuclear Receptors and their Families.Med Chem. 2020;16(5):594-604. doi: 10.2174/1573406415666191004125551. Med Chem. 2020. PMID: 31584374 Review.
-
iNuc-PhysChem: a sequence-based predictor for identifying nucleosomes via physicochemical properties.PLoS One. 2012;7(10):e47843. doi: 10.1371/journal.pone.0047843. Epub 2012 Oct 29. PLoS One. 2012. PMID: 23144709 Free PMC article.
-
iNR-Drug: predicting the interaction of drugs with nuclear receptors in cellular networking.Int J Mol Sci. 2014 Mar 19;15(3):4915-37. doi: 10.3390/ijms15034915. Int J Mol Sci. 2014. PMID: 24651462 Free PMC article.
Cited by
-
Comprehensive comparative analysis and identification of RNA-binding protein domains: multi-class classification and feature selection.J Theor Biol. 2012 Nov 7;312:65-75. doi: 10.1016/j.jtbi.2012.07.013. Epub 2012 Aug 3. J Theor Biol. 2012. PMID: 22884576 Free PMC article.
-
Accurate prediction of nuclear receptors with conjoint triad feature.BMC Bioinformatics. 2015 Dec 3;16:402. doi: 10.1186/s12859-015-0828-1. BMC Bioinformatics. 2015. PMID: 26630876 Free PMC article.
-
iCataly-PseAAC: Identification of Enzymes Catalytic Sites Using Sequence Evolution Information with Grey Model GM (2,1).J Membr Biol. 2015 Dec;248(6):1033-41. doi: 10.1007/s00232-015-9815-8. Epub 2015 Jun 16. J Membr Biol. 2015. PMID: 26077845
-
iDNA-Prot|dis: identifying DNA-binding proteins by incorporating amino acid distance-pairs and reduced alphabet profile into the general pseudo amino acid composition.PLoS One. 2014 Sep 3;9(9):e106691. doi: 10.1371/journal.pone.0106691. eCollection 2014. PLoS One. 2014. PMID: 25184541 Free PMC article.
-
PFP-GO: Integrating protein sequence, domain and protein-protein interaction information for protein function prediction using ranked GO terms.Front Genet. 2022 Sep 29;13:969915. doi: 10.3389/fgene.2022.969915. eCollection 2022. Front Genet. 2022. PMID: 36246645 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

