Analysis of the Tomato spotted wilt virus ambisense S RNA-encoded hairpin structure in translation
- PMID: 22363535
- PMCID: PMC3283609
- DOI: 10.1371/journal.pone.0031013
Analysis of the Tomato spotted wilt virus ambisense S RNA-encoded hairpin structure in translation
Abstract
Background: The intergenic region (IR) of ambisense RNA segments from animal- and plant-infecting (-)RNA viruses functions as a bidirectional transcription terminator. The IR sequence of the Tomato spotted wilt virus (TSWV) ambisense S RNA contains stretches that are highly rich in A-residues and U-residues and is predicted to fold into a stable hairpin structure. The presence of this hairpin structure sequence in the 3' untranslated region (UTR) of TSWV mRNAs implies a possible role in translation.
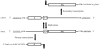
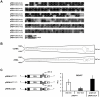
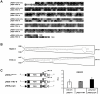
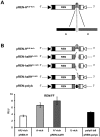
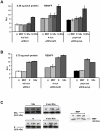
Methodology/principal findings: To analyse the role of the predicted hairpin structure in translation, various Renilla luciferase constructs containing modified 3' and/or 5' UTR sequences of the TSWV S RNA encoded nucleocapsid (N) gene were analyzed for expression. While good luciferase expression levels were obtained from constructs containing the 5' UTR and the 3' UTR, luciferase expression was lost when the hairpin structure sequence was removed from the 3' UTR. Constructs that only lacked the 5' UTR, still rendered good expression levels. When in addition the entire 3' UTR was exchanged for that of the S RNA encoded non-structural (NSs) gene transcript, containing the complementary hairpin folding sequence, the loss of luciferase expression could only be recovered by providing the 5' UTR sequence of the NSs transcript. Luciferase activity remained unaltered when the hairpin structure sequence was swapped for the analogous one from Tomato yellow ring virus, another distinct tospovirus. The addition of N and NSs proteins further increased luciferase expression levels from hairpin structure containing constructs.
Conclusions/significance: The results suggest a role for the predicted hairpin structure in translation in concert with the viral N and NSs proteins. The presence of stretches highly rich in A-residues does not rule out a concerted action with a poly(A)-tail-binding protein. A common transcription termination and translation strategy for plant- and animal-infecting ambisense RNA viruses is being discussed.
Conflict of interest statement
Figures
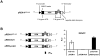
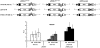

Similar articles
-
Analysis of the A-U rich hairpin from the intergenic region of tospovirus S RNA as target and inducer of RNA silencing.PLoS One. 2014 Sep 30;9(9):e106027. doi: 10.1371/journal.pone.0106027. eCollection 2014. PLoS One. 2014. PMID: 25268120 Free PMC article.
-
Tomato spotted wilt virus S-segment mRNAs have overlapping 3'-ends containing a predicted stem-loop structure and conserved sequence motif.Virus Res. 2005 Jun;110(1-2):125-31. doi: 10.1016/j.virusres.2005.01.012. Virus Res. 2005. PMID: 15845263
-
Variability of the N-protein and the intergenic region of the S RNA of tomato spotted wilt tospovirus (TSWV).New Microbiol. 2001 Apr;24(2):175-87. New Microbiol. 2001. PMID: 11346302
-
The amazing diversity of cap-independent translation elements in the 3'-untranslated regions of plant viral RNAs.Biochem Soc Trans. 2007 Dec;35(Pt 6):1629-33. doi: 10.1042/BST0351629. Biochem Soc Trans. 2007. PMID: 18031280 Free PMC article. Review.
-
A switch in time: detailing the life of a riboswitch.Biochim Biophys Acta. 2009 Sep-Oct;1789(9-10):584-91. doi: 10.1016/j.bbagrm.2009.06.004. Epub 2009 Jul 9. Biochim Biophys Acta. 2009. PMID: 19595806 Free PMC article. Review.
Cited by
-
Complementation between two tospoviruses facilitates the systemic movement of a plant virus silencing suppressor in an otherwise restrictive host.PLoS One. 2012;7(10):e44803. doi: 10.1371/journal.pone.0044803. Epub 2012 Oct 16. PLoS One. 2012. PMID: 23077485 Free PMC article.
-
Interplay between viruses and host mRNA degradation.Biochim Biophys Acta. 2013 Jun-Jul;1829(6-7):732-41. doi: 10.1016/j.bbagrm.2012.12.003. Epub 2012 Dec 26. Biochim Biophys Acta. 2013. PMID: 23274304 Free PMC article. Review.
-
Variation Profile of the Orthotospovirus Genome.Pathogens. 2020 Jun 29;9(7):521. doi: 10.3390/pathogens9070521. Pathogens. 2020. PMID: 32610472 Free PMC article.
-
Analysis of the A-U rich hairpin from the intergenic region of tospovirus S RNA as target and inducer of RNA silencing.PLoS One. 2014 Sep 30;9(9):e106027. doi: 10.1371/journal.pone.0106027. eCollection 2014. PLoS One. 2014. PMID: 25268120 Free PMC article.
-
Bunyaviral N Proteins Localize at RNA Processing Bodies and Stress Granules: The Enigma of Cytoplasmic Sources of Capped RNA for Cap Snatching.Viruses. 2022 Jul 29;14(8):1679. doi: 10.3390/v14081679. Viruses. 2022. PMID: 36016301 Free PMC article.
References
-
- Nguyen M, Haenni AL. Expression strategies of ambisense viruses. Vir Res. 2003;93:141–150. - PubMed
-
- Geerts-Dimitriadou C, Goldbach R, Kormelink R. Preferential use of RNA leader sequences during influenza A transcription initiation in vivo. Virology. 2011a;409:27–32. - PubMed
-
- Geerts-Dimitriadou C, Zwart MP, Goldbach R, Kormelink R. Base-pairing promotes leader selection to prime in vitro influenza genome transcription. Virology. 2011b;409:17–26. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

