Genome-wide analysis of the complex transcriptional networks of rice developing seeds
- PMID: 22363552
- PMCID: PMC3281924
- DOI: 10.1371/journal.pone.0031081
Genome-wide analysis of the complex transcriptional networks of rice developing seeds
Abstract
Background: The development of rice (Oryza sativa) seed is closely associated with assimilates storage and plant yield, and is fine controlled by complex regulatory networks. Exhaustive transcriptome analysis of developing rice embryo and endosperm will help to characterize the genes possibly involved in the regulation of seed development and provide clues of yield and quality improvement.
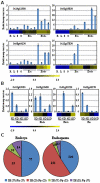
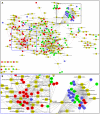
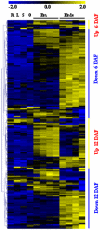
Principal findings: Our analysis showed that genes involved in metabolism regulation, hormone response and cellular organization processes are predominantly expressed during rice development. Interestingly, 191 transcription factor (TF)-encoding genes are predominantly expressed in seed and 59 TFs are regulated during seed development, some of which are homologs of seed-specific TFs or regulators of Arabidopsis seed development. Gene co-expression network analysis showed these TFs associated with multiple cellular and metabolism pathways, indicating a complex regulation of rice seed development. Further, by employing a cold-resistant cultivar Hanfeng (HF), genome-wide analyses of seed transcriptome at normal and low temperature reveal that rice seed is sensitive to low temperature at early stage and many genes associated with seed development are down-regulated by low temperature, indicating that the delayed development of rice seed by low temperature is mainly caused by the inhibition of the development-related genes. The transcriptional response of seed and seedling to low temperature is different, and the differential expressions of genes in signaling and metabolism pathways may contribute to the chilling tolerance of HF during seed development.
Conclusions: These results provide informative clues and will significantly improve the understanding of rice seed development regulation and the mechanism of cold response in rice seed.
Conflict of interest statement
Figures






References
-
- Itoh J, Nonomura K, Ikeda K, Yamaki S, Inukai Y, et al. Rice plant development: from zygote to spikelet. Plant Cell Physiol. 2005;46:23–47. - PubMed
-
- Sato Y, Tamaoki M, Murakami T, Yamamoto N, Kano-Murakami Y, et al. Abnormal cell divisions in leaf primordia caused by the expression of the rice homeobox gene OSH1 lead to altered morphology of leaves in transgenic tobacco. Mol Gen Genet. 1996;251:13–22. - PubMed
-
- Agarwal P, Kapoor S, Tyagi AK. Transcription factors regulating the progression of monocot and dicot seed development. Bioessays. 2011;33:189–202. - PubMed
-
- Kurata N, Miyoshi K, Nonomura K, Yamazaki Y, Ito Y. Rice mutants and genes related to organ development, morphogenesis and physiological traits. Plant Cell Physiol. 2005;46:48–62. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

