Highly sensitive detection of Staphylococcus aureus directly from patient blood
- PMID: 22363564
- PMCID: PMC3281916
- DOI: 10.1371/journal.pone.0031126
Highly sensitive detection of Staphylococcus aureus directly from patient blood
Abstract
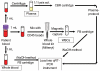
Background: Rapid detection of bloodstream infections (BSIs) can be lifesaving. We investigated the sample processing and assay parameters necessary for highly-sensitive detection of bloodstream bacteria, using Staphylococcus aureus as a model pathogen and an automated fluidic sample processing-polymerase chain reaction (PCR) platform as a model diagnostic system.
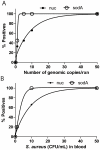
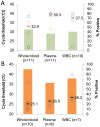
Methodology/principal findings: We compared a short 128 bp amplicon hemi-nested PCR and a relatively shorter 79 bp amplicon nested PCR targeting the S. aureus nuc and sodA genes, respectively. The sodA nested assay showed an enhanced limit of detection (LOD) of 5 genomic copies per reaction or 10 colony forming units (CFU) per ml blood over 50 copies per reaction or 50 CFU/ml for the nuc assay. To establish optimal extraction protocols, we investigated the relative abundance of the bacteria in different components of the blood (white blood cells (WBCs), plasma or whole blood), using the above assays. The blood samples were obtained from the patients who were culture positive for S. aureus. Whole blood resulted in maximum PCR positives with sodA assay (90% positive) as opposed to cell-associated bacteria (in WBCs) (71% samples positive) or free bacterial DNA in plasma (62.5% samples positive). Both the assays were further tested for direct detection of S. aureus in patient whole blood samples that were contemporaneous culture positive. S. aureus was detected in 40/45 of culture-positive patients (sensitivity 89%, 95% CI 0.75-0.96) and 0/59 negative controls with the sodA assay (specificity 100%, 95% CI 0.92-1).
Conclusions: We have demonstrated a highly sensitive two-hour assay for detection of sepsis causing bacteria like S. aureus directly in 1 ml of whole blood, without the need for blood culture.
Conflict of interest statement
Figures

Similar articles
-
Detection of Staphylococcus aureus by polymerase chain reaction amplification of the nuc gene.J Clin Microbiol. 1992 Jul;30(7):1654-60. doi: 10.1128/jcm.30.7.1654-1660.1992. J Clin Microbiol. 1992. PMID: 1629319 Free PMC article.
-
Evaluation of three different molecular markers for the detection of Staphylococcus aureus by polymerase chain reaction.Food Microbiol. 2008 May;25(3):452-9. doi: 10.1016/j.fm.2008.01.010. Epub 2008 Feb 2. Food Microbiol. 2008. PMID: 18355670
-
[A broad-range 16S rRNA gene real-time PCR assay for the diagnosis of neonatal septicemia].Zhonghua Er Ke Za Zhi. 2007 Jun;45(6):446-9. Zhonghua Er Ke Za Zhi. 2007. PMID: 17880793 Chinese.
-
Double triplex real-time PCR assay for simultaneous detection of Staphylococcus aureus, Staphylococcus epidermidis, Staphylococcus hominis, and Staphylococcus haemolyticus and determination of their methicillin resistance directly from positive blood culture bottles.Res Microbiol. 2011 Dec;162(10):1060-6. doi: 10.1016/j.resmic.2011.07.009. Epub 2011 Sep 3. Res Microbiol. 2011. PMID: 21925597
-
A culture-free biphasic approach for sensitive and rapid detection of pathogens in dried whole-blood matrix.Proc Natl Acad Sci U S A. 2022 Oct 4;119(40):e2209607119. doi: 10.1073/pnas.2209607119. Epub 2022 Sep 26. Proc Natl Acad Sci U S A. 2022. PMID: 36161889 Free PMC article.
Cited by
-
A 2020 review on the role of procalcitonin in different clinical settings: an update conducted with the tools of the Evidence Based Laboratory Medicine.Ann Transl Med. 2020 May;8(9):610. doi: 10.21037/atm-20-1855. Ann Transl Med. 2020. PMID: 32566636 Free PMC article. Review.
-
Sensitive Detection of Francisella tularensis Directly from Whole Blood by Use of the GeneXpert System.J Clin Microbiol. 2016 Dec 28;55(1):291-301. doi: 10.1128/JCM.01126-16. Print 2017 Jan. J Clin Microbiol. 2016. PMID: 27847371 Free PMC article.
-
PCR-based Approaches for the Detection of Clinical Methicillin-resistant Staphylococcus aureus.Open Microbiol J. 2016 Apr 14;10:45-56. doi: 10.2174/1874285801610010045. eCollection 2016. Open Microbiol J. 2016. PMID: 27335617 Free PMC article.
-
Advances in Directly Amplifying Nucleic Acids from Complex Samples.Biosensors (Basel). 2019 Sep 30;9(4):117. doi: 10.3390/bios9040117. Biosensors (Basel). 2019. PMID: 31574959 Free PMC article. Review.
-
The changing culture of the microbiology laboratory.Can J Infect Dis Med Microbiol. 2013 Fall;24(3):125-8. doi: 10.1155/2013/101630. Can J Infect Dis Med Microbiol. 2013. PMID: 24421822 Free PMC article. No abstract available.
References
-
- Gaibani P, Rossini G, Ambretti S, Gelsomino F, Pierro AM, et al. Blood culture systems: rapid detection - how and why? International Journal of Antimicrobial Agents. 2009;34:S13–S15. - PubMed
-
- Harbarth S, Sax H, Gastmeier P. The preventable proportion of nosocomial infections: an overview of published reports. Journal of Hospital Infection. 2003;54:258–266. - PubMed
-
- Kollef MH. The importance of appropriate initial antibiotic therapy for hospital-acquired infections. The American Journal of Medicine. 2003;115:582–584. - PubMed
-
- Lodise TP, McKinnon PS, Swiderski L, Rybak MJ. Outcomes Analysis of Delayed Antibiotic Treatment for Hospital-Acquired Staphylococcus aureus Bacteremia. Clinical Infectious Diseases. 2003;36:1418–1423. - PubMed
-
- Paolucci M, Capretti MG, Dal Monte P, Corvaglia L, Landini MP, et al. Laboratory diagnosis of late-onset sepsis in newborns by multiplex real-time PCR. Journal of Medical Microbiology. 2009;58:533–534. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

