New binding mode to TNF-alpha revealed by ubiquitin-based artificial binding protein
- PMID: 22363609
- PMCID: PMC3282696
- DOI: 10.1371/journal.pone.0031298
New binding mode to TNF-alpha revealed by ubiquitin-based artificial binding protein
Abstract
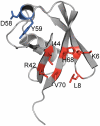
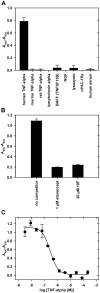
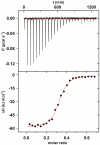
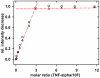
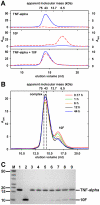
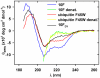
A variety of approaches have been employed to generate binding proteins from non-antibody scaffolds. Utilizing a beta-sheet of the human ubiquitin for paratope creation we obtained binding proteins against tumor necrosis factor (TNF)-alpha. The bioactive form of this validated pharmacological target protein is a non-covalently linked homo-trimer. This structural feature leads to the observation of a certain heterogeneity concerning the binding mode of TNF-alpha binding molecules, for instance in terms of monomer/trimer specificity. We analyzed a ubiquitin-based TNF-alpha binder, selected by ribosome display, with a particular focus on its mode of interaction. Using enzyme-linked immunosorbent assays, specific binding to TNF-alpha with nanomolar affinity was observed. In isothermal titration calorimetry we obtained comparable results regarding the affinity and detected an exothermic reaction with one ubiquitin-derived binding molecule binding one TNF-alpha trimer. Using NMR spectroscopy and other analytical methods the 1:3 stoichiometry could be confirmed. Detailed binding analysis showed that the interaction is affected by the detergent Tween-20. Previously, this phenomenon was reported only for one other type of alternative scaffold-derived binding proteins--designed ankyrin repeat proteins--without further investigation. As demonstrated by size exclusion chromatography and NMR spectroscopy, the presence of the detergent increases the association rate significantly. Since the special architecture of TNF-alpha is known to be modulated by detergents, the access to the recognized epitope is indicated to be restricted by conformational transitions within the target protein. Our results suggest that the ubiquitin-derived binding protein targets a new epitope on TNF-alpha, which differs from the epitopes recognized by TNF-alpha neutralizing antibodies.
Conflict of interest statement
Figures
Similar articles
-
Recombinant 55-kDa tumor necrosis factor (TNF) receptor. Stoichiometry of binding to TNF alpha and TNF beta and inhibition of TNF activity.J Biol Chem. 1991 Sep 25;266(27):18324-9. J Biol Chem. 1991. PMID: 1655744
-
Engineering bispecificity into a single albumin-binding domain.PLoS One. 2011;6(10):e25791. doi: 10.1371/journal.pone.0025791. Epub 2011 Oct 3. PLoS One. 2011. PMID: 21991353 Free PMC article.
-
Ubiquitin binding modulates IAP antagonist-stimulated proteasomal degradation of c-IAP1 and c-IAP2(1).Biochem J. 2009 Jan 1;417(1):149-60. doi: 10.1042/BJ20081885. Biochem J. 2009. PMID: 18939944
-
Structure-function relationship of tumor necrosis factor (TNF) and its receptor interaction based on 3D structural analysis of a fully active TNFR1-selective TNF mutant.J Mol Biol. 2009 Jan 30;385(4):1221-9. doi: 10.1016/j.jmb.2008.11.053. Epub 2008 Dec 6. J Mol Biol. 2009. PMID: 19084540
-
In vitro-engineered non-antibody protein therapeutics.Protein Cell. 2018 Jan;9(1):3-14. doi: 10.1007/s13238-017-0386-6. Epub 2017 Mar 7. Protein Cell. 2018. PMID: 28271446 Free PMC article. Review.
Cited by
-
A Structure-Based Strategy for Engineering Selective Ubiquitin Variant Inhibitors of Skp1-Cul1-F-Box Ubiquitin Ligases.Structure. 2018 Sep 4;26(9):1226-1236.e3. doi: 10.1016/j.str.2018.06.004. Epub 2018 Jul 19. Structure. 2018. PMID: 30033217 Free PMC article.
-
Tumor Necrosis Factor-α Trimer Disassembly and Inactivation via Peptide-Small Molecule Synergy.ACS Chem Biol. 2020 Aug 21;15(8):2116-2124. doi: 10.1021/acschembio.0c00313. Epub 2020 Jul 14. ACS Chem Biol. 2020. PMID: 32662976 Free PMC article.
-
Emerging drug development technologies targeting ubiquitination for cancer therapeutics.Pharmacol Ther. 2019 Jul;199:139-154. doi: 10.1016/j.pharmthera.2019.03.003. Epub 2019 Mar 7. Pharmacol Ther. 2019. PMID: 30851297 Free PMC article. Review.
-
Ubiquitin-derived artificial binding proteins targeting oncofetal fibronectin reveal scaffold plasticity by β-strand slippage.Commun Biol. 2024 Jul 27;7(1):907. doi: 10.1038/s42003-024-06569-9. Commun Biol. 2024. PMID: 39068227 Free PMC article.
-
Selection of specific protein binders for pre-defined targets from an optimized library of artificial helicoidal repeat proteins (alphaRep).PLoS One. 2013 Aug 27;8(8):e71512. doi: 10.1371/journal.pone.0071512. eCollection 2013. PLoS One. 2013. PMID: 24014183 Free PMC article.
References
-
- Glockshuber R, Malia M, Pfitzinger I, Plueckthun A. A comparison of strategies to stabilize immunoglobulin Fv-fragments. Biochemistry. 1990;29:1362–1367. - PubMed
-
- Hey T, Fiedler E, Rudolph R, Fiedler M. Artificial, non-antibody binding proteins for pharmaceutical and industrial applications. Trends Biotechnol. 2005;23:514–522. - PubMed
-
- Schlehuber S, Skerra A. Tuning ligand affinity, specificity, and folding stability of an engineered lipocalin variant - a so-called ‘anticalin’ - using a molecular random approach. Biophys Chem. 2002;96:213–228. - PubMed
-
- Nord K, Gunneriusson E, Ringdahl J, Stahl S, Uhlen M, et al. Binding proteins selected from combinatorial libraries of an [alpha]-helical bacterial receptor domain. Nat Biotech. 1997;15:772–777. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

