MLVA subtyping of genovar E Chlamydia trachomatis individualizes the Swedish variant and anorectal isolates from men who have sex with men
- PMID: 22363667
- PMCID: PMC3283677
- DOI: 10.1371/journal.pone.0031538
MLVA subtyping of genovar E Chlamydia trachomatis individualizes the Swedish variant and anorectal isolates from men who have sex with men
Abstract
This study describes a new multilocus variable number tandem-repeat (VNTR) analysis (MLVA) typing system for the discrimination of Chlamydia trachomatis genovar D to K isolates or specimens. We focused our MLVA scheme on genovar E which predominates in most populations worldwide. This system does not require culture and therefore can be performed directly on DNA extracted from positive clinical specimens. Our method was based on GeneScan analysis of five VNTR loci labelled with fluorescent dyes by multiplex PCR and capillary electrophoresis. This MLVA, called MLVA-5, was applied to a collection of 220 genovar E and 94 non-E genovar C. trachomatis isolates and specimens obtained from 251 patients and resulted in 38 MLVA-5 types. The genetic stability of the MLVA-5 scheme was assessed for results obtained both in vitro by serial passage culturing and in vivo using concomitant and sequential isolates and specimens. All anorectal genovar E isolates from men who have sex with men exhibited the same MLVA-5 type, suggesting clonal spread. In the same way, we confirmed the clonal origin of the Swedish new variant of C. trachomatis. The MLVA-5 assay was compared to three other molecular typing methods, ompA gene sequencing, multilocus sequence typing (MLST) and a previous MLVA method called MLVA-3, on 43 genovar E isolates. The discriminatory index was 0.913 for MLVA-5, 0.860 for MLST and 0.622 for MLVA-3. Among all of these genotyping methods, MLVA-5 displayed the highest discriminatory power and does not require a time-consuming sequencing step. The results indicate that MLVA-5 enables high-resolution molecular epidemiological characterisation of C. trachomatis genovars D to K infections directly from specimens.
Conflict of interest statement
Figures
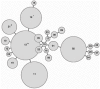

References
-
- Bébéar C, de Barbeyrac B. Genital Chlamydia trachomatis infections. Clin Microbiol Infect. 2009;15:4–10. - PubMed
-
- Ikryannikova LN, Shkarupeta MM, Shitikov EA, Il'ina EN, Govorun VM. Comparative evaluation of new typing schemes for urogenital Chlamydia trachomatis isolates. FEMS Immunol Med Microbiol. 2010;59:188–196. - PubMed
-
- Jurstrand M, Christerson L, Klint M, Fredlund H, Unemo M, et al. Characterisation of Chlamydia trachomatis by ompA sequencing and multilocus sequence typing in a Swedish county before and after identification of the new variant. Sex Transm Infect. 2010;86:56–60. - PubMed
-
- Mossman D, Beagley KW, Landay AL, Loewenthal M, Ooi C, et al. Genotyping of urogenital Chlamydia trachomatis in Regional New South Wales, Australia. Sex Transm Dis. 2008;35:614–616. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

