Transcriptional repressor NIR functions in the ribosome RNA processing of both 40S and 60S subunits
- PMID: 22363708
- PMCID: PMC3282729
- DOI: 10.1371/journal.pone.0031692
Transcriptional repressor NIR functions in the ribosome RNA processing of both 40S and 60S subunits
Abstract
Background: NIR was identified as an inhibitor of histone acetyltransferase and it represses transcriptional activation of p53. NIR is predominantly localized in the nucleolus and known as Noc2p, which is involved in the maturation of the 60S ribosomal subunit. However, how NIR functions in the nucleolus remains undetermined. In the nucleolus, a 47S ribosomal RNA precursor (pre-rRNA) is transcribed and processed to produce 18S, 5.8S and 28S rRNAs. The 18S rRNA is incorporated into the 40S ribosomal subunit, whereas the 28S and 5.8S rRNAs are incorporated into the 60S subunit. U3 small nucleolar RNA (snoRNA) directs 18S rRNA processing and U8 snoRNA mediates processing of 28S and 5.8 S rRNAs. Functional disruption of nucleolus often causes p53 activation to inhibit cell proliferation.
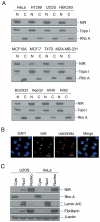
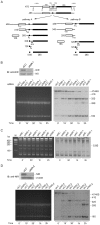
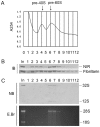
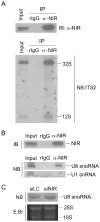
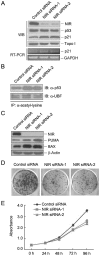
Methodology/principal findings: Western blotting showed that NIR is ubiquitously expressed in different human cell lines. Knock-down of NIR by siRNA led to inhibition of the 18S, 28S and 5.8S rRNAs evaluated by pulse-chase experiment. Pre-rRNA particles (pre-rRNPs) were fractionated from the nucleus by sucrose gradient centrifugation and analysis of the pre-RNPs components showed that NIR existed in the pre-RNPs of both the 60S and 40S subunits and co-fractionated with 32S and 12S pre-rRNAs in the 60S pre-rRNP. Protein-RNA binding experiments demonstrated that NIR is associated with the 32S pre-rRNA and U8 snoRNA. In addition, NIR bound U3 snoRNA. It is a novel finding that depletion of NIR did not affect p53 protein level but de-repressed acetylation of p53 and activated p21.
Conclusions: We provide the first evidence for a transcriptional repressor to function in the rRNA biogenesis of both the 40S and 60S subunits. Our findings also suggested that a nucleolar protein may alternatively signal to p53 by affecting the p53 modification rather than affecting p53 protein level.
Conflict of interest statement
Figures

Similar articles
-
Bop1 is a mouse WD40 repeat nucleolar protein involved in 28S and 5. 8S RRNA processing and 60S ribosome biogenesis.Mol Cell Biol. 2000 Aug;20(15):5516-28. doi: 10.1128/MCB.20.15.5516-5528.2000. Mol Cell Biol. 2000. PMID: 10891491 Free PMC article.
-
Splicing factor 2-associated protein p32 participates in ribosome biogenesis by regulating the binding of Nop52 and fibrillarin to preribosome particles.Mol Cell Proteomics. 2011 Aug;10(8):M110.006148. doi: 10.1074/mcp.M110.006148. Epub 2011 May 2. Mol Cell Proteomics. 2011. PMID: 21536856 Free PMC article.
-
Human nucleolar protein Nop52 (RRP1/NNP-1) is involved in site 2 cleavage in internal transcribed spacer 1 of pre-rRNAs at early stages of ribosome biogenesis.Nucleic Acids Res. 2015 Jun 23;43(11):5524-36. doi: 10.1093/nar/gkv470. Epub 2015 May 12. Nucleic Acids Res. 2015. PMID: 25969445 Free PMC article.
-
The multifunctional nucleolus.Nat Rev Mol Cell Biol. 2007 Jul;8(7):574-85. doi: 10.1038/nrm2184. Nat Rev Mol Cell Biol. 2007. PMID: 17519961 Review.
-
SUMO routes ribosome maturation.Nucleus. 2011 Nov-Dec;2(6):527-32. doi: 10.4161/nucl.2.6.17604. Epub 2011 Nov 1. Nucleus. 2011. PMID: 22064470 Review.
Cited by
-
A Novel Retinoblastoma Protein (RB) E3 Ubiquitin Ligase (NRBE3) Promotes RB Degradation and Is Transcriptionally Regulated by E2F1 Transcription Factor.J Biol Chem. 2015 Nov 20;290(47):28200-28213. doi: 10.1074/jbc.M115.655597. Epub 2015 Oct 6. J Biol Chem. 2015. PMID: 26442585 Free PMC article.
-
From Flies to Humans: A Conserved Role of CEBPZ, NOC2L, and NOC3L in rRNA Processing and Tumorigenesis.bioRxiv [Preprint]. 2025 Apr 23:2025.01.11.632529. doi: 10.1101/2025.01.11.632529. bioRxiv. 2025. Update in: J Cell Sci. 2025 Aug 19:jcs.264096. doi: 10.1242/jcs.264096. PMID: 39868138 Free PMC article. Updated. Preprint.
-
Unmasking the biological function and regulatory mechanism of NOC2L: a novel inhibitor of histone acetyltransferase.J Transl Med. 2023 Jan 17;21(1):31. doi: 10.1186/s12967-023-03877-2. J Transl Med. 2023. PMID: 36650543 Free PMC article. Review.
-
Rrp5p, Noc1p and Noc2p form a protein module which is part of early large ribosomal subunit precursors in S. cerevisiae.Nucleic Acids Res. 2013 Jan;41(2):1191-210. doi: 10.1093/nar/gks1056. Epub 2012 Dec 2. Nucleic Acids Res. 2013. PMID: 23209026 Free PMC article.
-
Cellular senescence and protein degradation: breaking down cancer.Cell Cycle. 2014;13(12):1840-58. doi: 10.4161/cc.29335. Epub 2014 May 27. Cell Cycle. 2014. PMID: 24866342 Free PMC article.
References
-
- Lapik YR, Fernandes CJ, Lau LF, Pestov DG. Physical and functional interaction between Pes1 and Bop1 in mammalian ribosome biogenesis. Mol Cell. 2004;15:17–29. - PubMed
-
- Balakin AG, Smith L, Fournier MJ. The RNA world of the nucleolus: two major families of small RNAs defined by different box elements with related functions. Cell. 1996;86:823–834. - PubMed
-
- Decatur WA, Fournier MJ. RNA-guided nucleotide modification of ribosomal and other RNAs. J Biol Chem. 2003;278:695–698. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

