Liver-specific expression of transcriptionally active SREBP-1c is associated with fatty liver and increased visceral fat mass
- PMID: 22363740
- PMCID: PMC3283692
- DOI: 10.1371/journal.pone.0031812
Liver-specific expression of transcriptionally active SREBP-1c is associated with fatty liver and increased visceral fat mass
Abstract
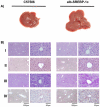
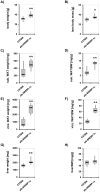
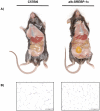
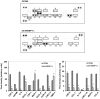
The pathogenesis of fatty liver is not understood in detail, but lipid overflow as well as de novo lipogenesis (DNL) seem to be the key points of hepatocyte accumulation of lipids. One key transcription factor in DNL is sterol regulatory element-binding protein (SREBP)-1c. We generated mice with liver-specific over-expression of mature human SREBP-1c under control of the albumin promoter and a liver-specific enhancer (alb-SREBP-1c) to analyze systemic perturbations caused by this distinct alteration. SREBP-1c targets specific genes and causes key enzymes in DNL and lipid metabolism to be up-regulated. The alb-SREBP-1c mice developed hepatic lipid accumulation featuring a fatty liver by the age of 24 weeks under normocaloric nutrition. On a molecular level, clinical parameters and lipid-profiles varied according to the fatty liver phenotype. The desaturation index was increased compared to wild type mice. In liver, fatty acids (FA) were increased by 50% (p<0.01) and lipid composition was shifted to mono unsaturated FA, whereas lipid profile in adipose tissue or serum was not altered. Serum analyses revealed a ∼2-fold (p<0.01) increase in triglycerides and free fatty acids, and a ∼3-fold (p<0.01) increase in insulin levels, indicating insulin resistance; however, no significant cytokine profile alterations have been determined. Interestingly and unexpectedly, mice also developed adipositas with considerably increased visceral adipose tissue, although calorie intake was not different compared to control mice. In conclusion, the alb-SREBP-1c mouse model allowed the elucidation of the systemic impact of SREBP-1c as a central regulator of lipid metabolism in vivo and also demonstrated that the liver is a more active player in metabolic diseases such as visceral obesity and insulin resistance.
Conflict of interest statement
Figures


References
-
- Unger RH. Lipotoxic diseases. Annu Rev Med. 2002;53:19–36. - PubMed
-
- Choi SS, Diehl AM. Hepatic triglyceride synthesis and nonalcoholic fatty liver disease. Curr Opin Lipidol. 2008;19:295–300. - PubMed
-
- Musso G, Gambino R, Cassader M. Recent insights into hepatic lipid metabolism in non-alcoholic fatty liver disease (NAFLD). Prog Lipid. 2009;48:1–26. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

