Genomic scan of selective sweeps in thin and fat tail sheep breeds for identifying of candidate regions associated with fat deposition
- PMID: 22364287
- PMCID: PMC3351017
- DOI: 10.1186/1471-2156-13-10
Genomic scan of selective sweeps in thin and fat tail sheep breeds for identifying of candidate regions associated with fat deposition
Abstract
Background: Identification of genomic regions that have been targets of selection for phenotypic traits is one of the most important and challenging areas of research in animal genetics. However, currently there are relatively few genomic regions identified that have been subject to positive selection. In this study, a genome-wide scan using ~50,000 Single Nucleotide Polymorphisms (SNPs) was performed in an attempt to identify genomic regions associated with fat deposition in fat-tail breeds. This trait and its modification are very important in those countries grazing these breeds.
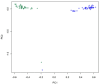
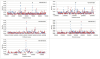
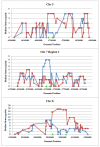
Results: Two independent experiments using either Iranian or Ovine HapMap genotyping data contrasted thin and fat tail breeds. Population differentiation using FST in Iranian thin and fat tail breeds revealed seven genomic regions. Almost all of these regions overlapped with QTLs that had previously been identified as affecting fat and carcass yield traits in beef and dairy cattle. Study of selection sweep signatures using FST in thin and fat tail breeds sampled from the Ovine HapMap project confirmed three of these regions located on Chromosomes 5, 7 and X. We found increased homozygosity in these regions in favour of fat tail breeds on chromosome 5 and X and in favour of thin tail breeds on chromosome 7.
Conclusions: In this study, we were able to identify three novel regions associated with fat deposition in thin and fat tail sheep breeds. Two of these were associated with an increase of homozygosity in the fat tail breeds which would be consistent with selection for mutations affecting fat tail size several thousand years after domestication.
Figures

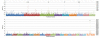
References
-
- Ryder ML. Sheep and Man. London: Duckworth press; 1983. pp. 17–92.
-
- Zeder MA. Animal domestication in the Zagros: a review of past and current research. Pale'orient. 1999;25:11–26.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

