Catalytic contributions of key residues in the adenine glycosylase MutY revealed by pH-dependent kinetics and cellular repair assays
- PMID: 22365610
- PMCID: PMC3292766
- DOI: 10.1016/j.chembiol.2011.11.011
Catalytic contributions of key residues in the adenine glycosylase MutY revealed by pH-dependent kinetics and cellular repair assays
Abstract
MutY prevent DNA mutations associated with 8-oxoguanine (OG) by catalyzing the removal of adenines opposite OG. pH dependence of the adenine glycosylase activity establish that Asp 138 of MutY must be deprotonated for maximal activity consistent with its role in stabilizing the oxacarbenium ion transition state in an S(N)1 mechanism. A cellular OG:A repair assay allowed further validation of the critical role of Asp 138. Conservative substitutions of the catalytic residues Asp 138 and Glu 37 resulted in enzymes with a range of activity that were used to correlate the efficiency of adenine excision with overall OG:A repair and suppression of DNA mutations in vivo. The results show that MutY variations that exhibit reduced mismatch affinity result in more dramatic reductions in cellular OG:A repair than those that only compromise adenine excision catalysis.
Copyright © 2012 Elsevier Ltd. All rights reserved.
Figures
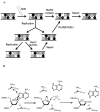
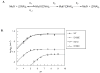


Comment in
-
For MutY, it's all about the OG.Chem Biol. 2012 Mar 23;19(3):313-4. doi: 10.1016/j.chembiol.2012.03.002. Chem Biol. 2012. PMID: 22444586 Free PMC article.
References
-
- Hoeijmakers JHJ. Genome Maitenance mechanisms for preventing cancers. Nature. 2001;411:366–374. - PubMed
-
- Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. ASM Press; Washington DC: 2006.
-
- David SS, Williams SD. Chemistry of Glycosylases and Endonucleases Involved in Base-Excision Repair. Chem Rev. 1998;98:1221–1261. - PubMed
-
- Burrows CM, Muller J. Oxidative Nucleobase Modifications Leading to Stand Scission. Chem Reviews. 1998;98:1109–1151. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

