Interplay between gene expression noise and regulatory network architecture
- PMID: 22365642
- PMCID: PMC3340541
- DOI: 10.1016/j.tig.2012.01.006
Interplay between gene expression noise and regulatory network architecture
Abstract
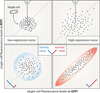
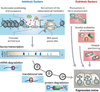
Complex regulatory networks orchestrate most cellular processes in biological systems. Genes in such networks are subject to expression noise, resulting in isogenic cell populations exhibiting cell-to-cell variation in protein levels. Increasing evidence suggests that cells have evolved regulatory strategies to limit, tolerate or amplify expression noise. In this context, fundamental questions arise: how can the architecture of gene regulatory networks generate, make use of or be constrained by expression noise? Here, we discuss the interplay between expression noise and gene regulatory network at different levels of organization, ranging from a single regulatory interaction to entire regulatory networks. We then consider how this interplay impacts a variety of phenomena, such as pathogenicity, disease, adaptation to changing environments, differential cell-fate outcome and incomplete or partial penetrance effects. Finally, we highlight recent technological developments that permit measurements at the single-cell level, and discuss directions for future research.
Copyright © 2012 Elsevier Ltd. All rights reserved.
Figures


References
-
- Spudich JL, Koshland DE. Non-genetic individuality: chance in the single cell. Nature. 1976;262:467–471. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

