Cellular noise regulons underlie fluctuations in Saccharomyces cerevisiae
- PMID: 22365828
- PMCID: PMC3327736
- DOI: 10.1016/j.molcel.2011.11.035
Cellular noise regulons underlie fluctuations in Saccharomyces cerevisiae
Abstract
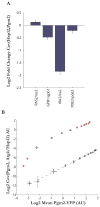
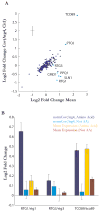
Stochasticity is a hallmark of cellular processes, and different classes of genes show large differences in their cell-to-cell variability (noise). To decipher the sources and consequences of this noise, we systematically measured pairwise correlations between large numbers of genes, including those with high variability. We find that there is substantial pathway variability shared across similarly regulated genes. This induces quantitative correlations in the expression of functionally related genes such as those involved in the Msn2/4 stress response pathway, amino-acid biosynthesis, and mitochondrial maintenance. Bioinformatic analyses and genetic perturbations suggest that fluctuations in PKA and Tor signaling contribute to pathway-specific variability. Our results argue that a limited number of well-delineated "noise regulons" operate across a yeast cell and that such coordinated fluctuations enable a stochastic but coherent induction of functionally related genes. Finally, we show that pathway noise is a quantitative tool for exploring pathway features and regulatory relationships in un-stimulated systems.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures
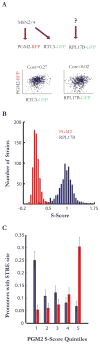
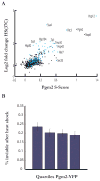
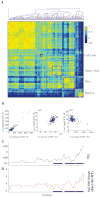
Comment in
-
When noisy neighbors are a blessing: analysis of gene expression noise identifies coregulated genes.Mol Cell. 2012 Feb 24;45(4):437-8. doi: 10.1016/j.molcel.2012.02.002. Mol Cell. 2012. PMID: 22365826
References
-
- Becskei A, Kaufmann BB, Oudenaarden A. Contributions of low molecular number and chromosomal positioning to stochastic gene expression. Nature Genetics. 2005;37:937–944. - PubMed
-
- Bigger W. Treatment Of Staphylococcal Infections With Penicillin by Intermittent Sterilisation. The Lancet. 1944;244(6320):497–500.
-
- Blake WJ, Kaern M, Cantor CR, Collins JJ. Noise in eukaryotic gene expression. Nature. 2003;422:633–637. - PubMed
-
- Blake WJ, Balazsi G, Kohanski MA, Isaacs FJ, Murphy KF, Kuang Y, Cantor CR, Walt DR, Collins JJ. Phenotypic consequences of promoter-mediated transcriptional noise. Molecular Cell. 2006;24:853–865. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

