Role of ATG8 and autophagy in programmed nuclear degradation in Tetrahymena thermophila
- PMID: 22366125
- PMCID: PMC3318292
- DOI: 10.1128/EC.05296-11
Role of ATG8 and autophagy in programmed nuclear degradation in Tetrahymena thermophila
Abstract
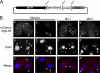
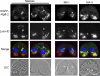
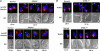
Autophagy is an evolutionarily conserved mechanism for the degradation of cellular components, but its role in enucleation during differentiation has not been established. Tetrahymena thermophila is a unicellular eukaryote with two functionally distinct nuclei, the somatic (macro-) and the germ line (micro-) nuclei. These nuclei are produced during sexual reproduction (conjugation), which involves differentiation and selective degradation of several specific nuclei. To examine the role of autophagy in nuclear degradation, we studied the function of two ATG8 genes in Tetrahymena. Through fluorescent protein tagging, we found that both proteins are targeted to degrading nuclei at specific stages, with some enrichment on the nuclear periphery, suggesting the formation of autophagosomes surrounding these nuclei. In addition, ATG8 knockout mutant cells showed a pronounced delay in nuclear degradation without apparently preventing the completion of other developmental events. This evidence provided direct support for a critical role for autophagy in programmed nuclear degradation. The results also showed differential roles for two ATG8 genes, with ATG8-65 playing a more significant role in starvation than ATG8-2, although both are important in nuclear degradation.
Figures




Similar articles
-
Atg5 Regulates Selective Autophagy of the Parental Macronucleus during Tetrahymena Sexual Reproduction.Cells. 2021 Nov 8;10(11):3071. doi: 10.3390/cells10113071. Cells. 2021. PMID: 34831293 Free PMC article.
-
Zygotic expression of the double-stranded RNA binding motif protein Drb2p is required for DNA elimination in the ciliate Tetrahymena thermophila.Eukaryot Cell. 2011 Dec;10(12):1648-59. doi: 10.1128/EC.05216-11. Epub 2011 Oct 21. Eukaryot Cell. 2011. PMID: 22021239 Free PMC article.
-
Conserved Asf1-importin β physical interaction in growth and sexual development in the ciliate Tetrahymena thermophila.J Proteomics. 2013 Dec 6;94:311-26. doi: 10.1016/j.jprot.2013.09.018. Epub 2013 Oct 11. J Proteomics. 2013. PMID: 24120531
-
Developmental progression of Tetrahymena through the cell cycle and conjugation.Methods Cell Biol. 2012;109:177-236. doi: 10.1016/B978-0-12-385967-9.00007-4. Methods Cell Biol. 2012. PMID: 22444146 Review.
-
Keeping the soma free of transposons: programmed DNA elimination in ciliates.J Biol Chem. 2011 Oct 28;286(43):37045-52. doi: 10.1074/jbc.R111.276964. Epub 2011 Sep 13. J Biol Chem. 2011. PMID: 21914793 Free PMC article. Review.
Cited by
-
Mechanistic insights into an atypical interaction between ATG8 and SH3P2 in Arabidopsis thaliana.Autophagy. 2022 Jun;18(6):1350-1366. doi: 10.1080/15548627.2021.1976965. Epub 2021 Oct 17. Autophagy. 2022. PMID: 34657568 Free PMC article.
-
The Transmembrane Protein Semi1 Positions Gamete Nuclei for Reciprocal Fertilization in Tetrahymena.iScience. 2020 Jan 24;23(1):100749. doi: 10.1016/j.isci.2019.100749. Epub 2019 Nov 28. iScience. 2020. PMID: 31884169 Free PMC article.
-
The opposing roles of the mTOR signaling pathway in different phases of human umbilical cord blood-derived CD34+ cell erythropoiesis.Stem Cells. 2020 Nov;38(11):1492-1505. doi: 10.1002/stem.3268. Epub 2020 Sep 1. Stem Cells. 2020. PMID: 32871057 Free PMC article.
-
Snf2 Proteins Are Required to Generate Gamete Pronuclei in Tetrahymena thermophila.Microorganisms. 2022 Dec 7;10(12):2426. doi: 10.3390/microorganisms10122426. Microorganisms. 2022. PMID: 36557679 Free PMC article.
-
Diversification of CORVET tethers facilitates transport complexity in Tetrahymena thermophila.J Cell Sci. 2020 Feb 12;133(3):jcs238659. doi: 10.1242/jcs.238659. J Cell Sci. 2020. PMID: 31964712 Free PMC article.
References
-
- Allewell NM, Oles J, Wolfe J. 1976. A physicochemical analysis of conjugation in Tetrahymena pyriformis. Exp. Cell Res. 97:394–405 - PubMed
-
- Alvarez VE, et al. 2008. Autophagy is involved in nutritional stress response and differentiation in Trypanosoma cruzi. J. Biol. Chem. 283:3454–3464 - PubMed
-
- Benchimol M. 1999. Hydrogenosome autophagy: an ultrastructural and cytochemical study. Biol. Cell 91:165–174 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

