STAY-GREEN and chlorophyll catabolic enzymes interact at light-harvesting complex II for chlorophyll detoxification during leaf senescence in Arabidopsis
- PMID: 22366162
- PMCID: PMC3315229
- DOI: 10.1105/tpc.111.089474
STAY-GREEN and chlorophyll catabolic enzymes interact at light-harvesting complex II for chlorophyll detoxification during leaf senescence in Arabidopsis
Abstract
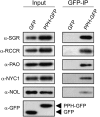
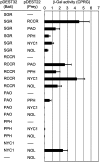
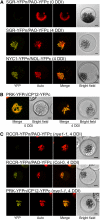
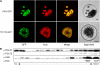
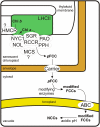
During leaf senescence, plants degrade chlorophyll to colorless linear tetrapyrroles that are stored in the vacuole of senescing cells. The early steps of chlorophyll breakdown occur in plastids. To date, five chlorophyll catabolic enzymes (CCEs), NONYELLOW COLORING1 (NYC1), NYC1-LIKE, pheophytinase, pheophorbide a oxygenase (PAO), and red chlorophyll catabolite reductase, have been identified; these enzymes catalyze the stepwise degradation of chlorophyll to a fluorescent intermediate, pFCC, which is then exported from the plastid. In addition, STAY-GREEN (SGR), Mendel's green cotyledon gene encoding a chloroplast protein, is required for the initiation of chlorophyll breakdown in plastids. Senescence-induced SGR binds to light-harvesting complex II (LHCII), but its exact role remains elusive. Here, we show that all five CCEs also specifically interact with LHCII. In addition, SGR and CCEs interact directly or indirectly with each other at LHCII, and SGR is essential for recruiting CCEs in senescing chloroplasts. PAO, which had been attributed to the inner envelope, is found to localize in the thylakoid membrane. These data indicate a predominant role for the SGR-CCE-LHCII protein interaction in the breakdown of LHCII-located chlorophyll, likely to allow metabolic channeling of phototoxic chlorophyll breakdown intermediates upstream of nontoxic pFCC.
Figures



Similar articles
-
7-Hydroxymethyl chlorophyll a reductase functions in metabolic channeling of chlorophyll breakdown intermediates during leaf senescence.Biochem Biophys Res Commun. 2013 Jan 4;430(1):32-7. doi: 10.1016/j.bbrc.2012.11.050. Epub 2012 Nov 27. Biochem Biophys Res Commun. 2013. PMID: 23200839
-
Arabidopsis STAY-GREEN2 is a negative regulator of chlorophyll degradation during leaf senescence.Mol Plant. 2014 Aug;7(8):1288-1302. doi: 10.1093/mp/ssu045. Epub 2014 Apr 9. Mol Plant. 2014. PMID: 24719469
-
The Stay-Green Rice like (SGRL) gene regulates chlorophyll degradation in rice.J Plant Physiol. 2013 Oct 15;170(15):1367-73. doi: 10.1016/j.jplph.2013.05.016. Epub 2013 Jun 28. J Plant Physiol. 2013. PMID: 23816327
-
What stay-green mutants tell us about nitrogen remobilization in leaf senescence.J Exp Bot. 2002 Apr;53(370):801-8. doi: 10.1093/jexbot/53.370.801. J Exp Bot. 2002. PMID: 11912223 Review.
-
The Divergent Roles of STAYGREEN (SGR) Homologs in Chlorophyll Degradation.Mol Cells. 2015 May;38(5):390-5. doi: 10.14348/molcells.2015.0039. Epub 2015 Apr 24. Mol Cells. 2015. PMID: 25913011 Free PMC article. Review.
Cited by
-
Stress-induced chloroplast degradation in Arabidopsis is regulated via a process independent of autophagy and senescence-associated vacuoles.Plant Cell. 2014 Dec;26(12):4875-88. doi: 10.1105/tpc.114.133116. Epub 2014 Dec 23. Plant Cell. 2014. PMID: 25538186 Free PMC article.
-
A Role for TIC55 as a Hydroxylase of Phyllobilins, the Products of Chlorophyll Breakdown during Plant Senescence.Plant Cell. 2016 Oct;28(10):2510-2527. doi: 10.1105/tpc.16.00630. Epub 2016 Sep 21. Plant Cell. 2016. PMID: 27655840 Free PMC article.
-
Update on the biochemistry of chlorophyll breakdown.Plant Mol Biol. 2013 Aug;82(6):505-17. doi: 10.1007/s11103-012-9940-z. Epub 2012 Jul 13. Plant Mol Biol. 2013. PMID: 22790503 Review.
-
Pheophorbide a May Regulate Jasmonate Signaling during Dark-Induced Senescence.Plant Physiol. 2020 Feb;182(2):776-791. doi: 10.1104/pp.19.01115. Epub 2019 Nov 21. Plant Physiol. 2020. PMID: 31753845 Free PMC article.
-
Rice 7-Hydroxymethyl Chlorophyll a Reductase Is Involved in the Promotion of Chlorophyll Degradation and Modulates Cell Death Signaling.Mol Cells. 2017 Oct;40(10):773-786. doi: 10.14348/molcells.2017.0127. Epub 2017 Oct 17. Mol Cells. 2017. PMID: 29047257 Free PMC article.
References
-
- Aubry S., Mani J., Hörtensteiner S. (2008). Stay-green protein, defective in Mendel’s green cotyledon mutant, acts independent and upstream of pheophorbide a oxygenase in the chlorophyll catabolic pathway. Plant Mol. Biol. 67: 243–256 - PubMed
-
- Bracha-Drori K., Shichrur K., Katz A., Oliva M., Angelovici R., Yalovsky S., Ohad N. (2004). Detection of protein-protein interactions in plants using bimolecular fluorescence complementation. Plant J. 40: 419–427 - PubMed
-
- Earley K.W., Haag J.R., Pontes O., Opper K., Juehne T., Song K.M., Pikaard C.S. (2006). Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45: 616–629 - PubMed
-
- Endler A., Meyer S., Schelbert S., Schneider T., Weschke W., Peters S.W., Keller F., Baginsky S., Martinoia E., Schmidt U.G. (2006). Identification of a vacuolar sucrose transporter in barley and Arabidopsis mesophyll cells by a tonoplast proteomic approach. Plant Physiol. 141: 196–207 - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

