Comparative analysis of the first complete Enterococcus faecium genome
- PMID: 22366422
- PMCID: PMC3347086
- DOI: 10.1128/JB.00259-12
Comparative analysis of the first complete Enterococcus faecium genome
Abstract
Vancomycin-resistant enterococci (VRE) are one of the leading causes of nosocomial infections in health care facilities around the globe. In particular, infections caused by vancomycin-resistant Enterococcus faecium are becoming increasingly common. Comparative and functional genomic studies of E. faecium isolates have so far been limited owing to the lack of a fully assembled E. faecium genome sequence. Here we address this issue and report the complete 3.0-Mb genome sequence of the multilocus sequence type 17 vancomycin-resistant Enterococcus faecium strain Aus0004, isolated from the bloodstream of a patient in Melbourne, Australia, in 1998. The genome comprises a 2.9-Mb circular chromosome and three circular plasmids. The chromosome harbors putative E. faecium virulence factors such as enterococcal surface protein, hemolysin, and collagen-binding adhesin. Aus0004 has a very large accessory genome (38%) that includes three prophage and two genomic islands absent among 22 other E. faecium genomes. One of the prophage was present as inverted 50-kb repeats that appear to have facilitated a 683-kb chromosomal inversion across the replication terminus, resulting in a striking replichore imbalance. Other distinctive features include 76 insertion sequence elements and a single chromosomal copy of Tn1549 containing the vanB vancomycin resistance element. A complete E. faecium genome will be a useful resource to assist our understanding of this emerging nosocomial pathogen.
Figures
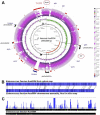
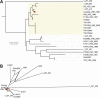
References
-
- Arias CA, et al. 2007. Failure of daptomycin monotherapy for endocarditis caused by an Enterococcus faecium strain with vancomycin-resistant and vancomycin-susceptible subpopulations and evidence of in vivo loss of the vanA gene cluster. Clin. Infect. Dis. 45:1343–1346 - PubMed
-
- Arthur M, Reynolds P, Courvalin P. 1996. Glycopeptide resistance in enterococci. Trends Microbiol. 4:401–407 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

