Sculpting the endomembrane system in deep time: high resolution phylogenetics of Rab GTPases
- PMID: 22366452
- PMCID: PMC3383260
- DOI: 10.1242/jcs.101378
Sculpting the endomembrane system in deep time: high resolution phylogenetics of Rab GTPases
Abstract
The presence of a nucleus and other membrane-bounded intracellular compartments is the defining feature of eukaryotic cells. Endosymbiosis accounts for the origins of mitochondria and plastids, but the evolutionary ancestry of the remaining cellular compartments is incompletely documented. Resolving the evolutionary history of organelle-identity encoding proteins within the endomembrane system is a necessity for unravelling the origins and diversification of the endogenously derived organelles. Comparative genomics reveals events after the last eukaryotic common ancestor (LECA), but resolution of events prior to LECA, and a full account of the intracellular compartments present in LECA, has proved elusive. We have devised and exploited a new phylogenetic strategy to reconstruct the history of the Rab GTPases, a key family of endomembrane-specificity proteins. Strikingly, we infer a remarkably sophisticated organellar composition for LECA, which we predict possessed as many as 23 Rab GTPases. This repertoire is significantly greater than that present in many modern organisms and unexpectedly indicates a major role for secondary loss in the evolutionary diversification of the endomembrane system. We have identified two Rab paralogues of unknown function but wide distribution, and thus presumably ancient nature; RabTitan and RTW. Furthermore, we show that many Rab paralogues emerged relatively suddenly during early metazoan evolution, which is in stark contrast to the lack of significant Rab family expansions at the onset of most other major eukaryotic groups. Finally, we reconstruct higher-order ancestral clades of Rabs primarily linked with endocytic and exocytic process, suggesting the presence of primordial Rabs associated with the establishment of those pathways and giving the deepest glimpse to date into pre-LECA history of the endomembrane system.
Figures
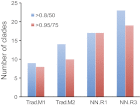
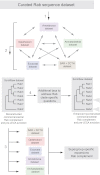
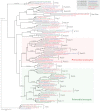


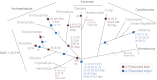
References
-
- Abascal F., Zardoya R., Posada D. (2005). ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21, 2104-2105 - PubMed
-
- Adl S. M., Simpson A. G., Farmer M. A., Andersen R. A., Anderson O. R., Barta J. R., Bowser S. S., Brugerolle G., Fensome R. A., Fredericq S., et al. (2005). The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. J. Eukaryot. Microbiol. 52, 399-451 - PubMed
-
- Ayong L., Pagnotti G., Tobon A. B., Chakrabarti D. (2007). Identification of Plasmodium falciparum family of SNAREs. Mol. Biochem. Parasitol. 152, 113-122 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

