Blind prediction of host-guest binding affinities: a new SAMPL3 challenge
- PMID: 22366955
- PMCID: PMC3383923
- DOI: 10.1007/s10822-012-9554-1
Blind prediction of host-guest binding affinities: a new SAMPL3 challenge
Abstract
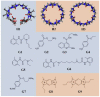
The computational prediction of protein-ligand binding affinities is of central interest in early-stage drug-discovery, and there is a widely recognized need for improved methods. Low molecular weight receptors and their ligands--i.e., host-guest systems--represent valuable test-beds for such affinity prediction methods, because their small size makes for fast calculations and relatively facile numerical convergence. The SAMPL3 community exercise included the first ever blind prediction challenge for host-guest binding affinities, through the incorporation of 11 new host-guest complexes. Ten participating research groups addressed this challenge with a variety of approaches. Statistical assessment indicates that, although most methods performed well at predicting some general trends in binding affinity, overall accuracy was not high, as all the methods suffered from either poor correlation or high RMS errors or both. There was no clear advantage in using explicit versus implicit solvent models, any particular force field, or any particular approach to conformational sampling. In a few cases, predictions using very similar energy models but different sampling and/or free-energy methods resulted in significantly different results. The protonation states of one host and some guest molecules emerged as key uncertainties beyond the choice of computational approach. The present results have implications for methods development and future blind prediction exercises.
Figures
Similar articles
-
Overview of the SAMPL5 host-guest challenge: Are we doing better?J Comput Aided Mol Des. 2017 Jan;31(1):1-19. doi: 10.1007/s10822-016-9974-4. Epub 2016 Sep 22. J Comput Aided Mol Des. 2017. PMID: 27658802 Free PMC article. Review.
-
Overview of the SAMPL6 host-guest binding affinity prediction challenge.J Comput Aided Mol Des. 2018 Oct;32(10):937-963. doi: 10.1007/s10822-018-0170-6. Epub 2018 Nov 10. J Comput Aided Mol Des. 2018. PMID: 30415285 Free PMC article.
-
The SAMPL4 host-guest blind prediction challenge: an overview.J Comput Aided Mol Des. 2014 Apr;28(4):305-17. doi: 10.1007/s10822-014-9735-1. Epub 2014 Mar 6. J Comput Aided Mol Des. 2014. PMID: 24599514 Free PMC article. Review.
-
SAMPL7 Host-Guest Challenge Overview: assessing the reliability of polarizable and non-polarizable methods for binding free energy calculations.J Comput Aided Mol Des. 2021 Jan;35(1):1-35. doi: 10.1007/s10822-020-00363-5. Epub 2021 Jan 4. J Comput Aided Mol Des. 2021. PMID: 33392951 Free PMC article.
-
Prediction of SAMPL3 host-guest affinities with the binding energy distribution analysis method (BEDAM).J Comput Aided Mol Des. 2012 May;26(5):505-16. doi: 10.1007/s10822-012-9552-3. Epub 2012 Feb 22. J Comput Aided Mol Des. 2012. PMID: 22354755 Free PMC article.
Cited by
-
Overview of the SAMPL5 host-guest challenge: Are we doing better?J Comput Aided Mol Des. 2017 Jan;31(1):1-19. doi: 10.1007/s10822-016-9974-4. Epub 2016 Sep 22. J Comput Aided Mol Des. 2017. PMID: 27658802 Free PMC article. Review.
-
Binding free energies in the SAMPL5 octa-acid host-guest challenge calculated with DFT-D3 and CCSD(T).J Comput Aided Mol Des. 2017 Jan;31(1):87-106. doi: 10.1007/s10822-016-9957-5. Epub 2016 Sep 6. J Comput Aided Mol Des. 2017. PMID: 27600554 Free PMC article.
-
Computational Calorimetry: High-Precision Calculation of Host-Guest Binding Thermodynamics.J Chem Theory Comput. 2015 Sep 8;11(9):4377-94. doi: 10.1021/acs.jctc.5b00405. J Chem Theory Comput. 2015. PMID: 26523125 Free PMC article.
-
The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities.Expert Opin Drug Discov. 2015 May;10(5):449-61. doi: 10.1517/17460441.2015.1032936. Epub 2015 Apr 2. Expert Opin Drug Discov. 2015. PMID: 25835573 Free PMC article. Review.
-
Detailed potential of mean force studies on host-guest systems from the SAMPL6 challenge.J Comput Aided Mol Des. 2018 Oct;32(10):1013-1026. doi: 10.1007/s10822-018-0153-7. Epub 2018 Aug 24. J Comput Aided Mol Des. 2018. PMID: 30143917
References
-
- Kitchen DB, Decornez H, Furr JR, Bajorath J. Docking and scoring in virtual screening for drug discovery: Methods and applications. Nat Rev Drug Discov. 2004;3:935–949. - PubMed
-
- Gilson MK, Zhou HX. Calculation of protein-ligand binding affinities. Annu Rev Bioph Biom. 2007;36:21–42. - PubMed
-
- Houk KN, Leach AG, Kim SP, Zhang X. Binding affinities of host-guest, protein-ligand, and protein-transition-state complexes. Angew Chem Int Ed Engl. 2003;42:4872–4897. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Chemical Information

