Genome scans for transmission ratio distortion regions in mice
- PMID: 22367040
- PMCID: PMC3338264
- DOI: 10.1534/genetics.111.135988
Genome scans for transmission ratio distortion regions in mice
Abstract
Transmission ratio distortion (TRD) is the departure from the expected genotypic frequencies under Mendelian inheritance. This departure can be due to multiple physiological mechanisms during gametogenesis, fertilization, fetal and embryonic development, and early neonatal life. Although a few TRD loci have been reported in mouse, inheritance patterns have never been evaluated for TRD. In this article, we developed a Bayesian binomial model accounting for additive and dominant deviation TRD mechanisms. Moreover, this model was used to perform genome-wide scans for TRD quantitative trait loci (QTL) on six F2 mouse crosses involving between 296 and 541 mice and between 72 and 1854 genetic markers. Statistical significance of each model was checked at each genetic marker with Bayes factors. Genome scans revealed overdominance TRD QTL located in mouse chromosomes 1, 2, 12, 13, and 14 and additive TRD QTL in mouse chromosomes 2, 3, and 15, although these results did not replicate across mouse crosses. This research contributes new statistical tools for the analysis of specific genetic patterns involved in TRD in F2 populations, our results suggesting a relevant incidence of TRD phenomena in mouse with important implications for both statistical analyses and biological research.
Figures


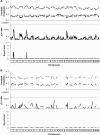
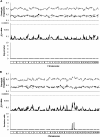
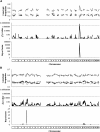
References
-
- Agulnik S. I., Agulnik A. I., Ruvinsky A. O., 1990. Meiotic drive in female mice heterozygous for the HSR insert on chromosome 1. Genet. Res. 55: 97–100. - PubMed
-
- Bonferroni C. E., 1930. Elementi di Statistica Generale. Libreria Seber, Florence, Italy.
-
- Bruce A. B., 1910. The Mendelian theory of heredity and the augmentation of vigor. Science 32: 627–628. - PubMed
-
- Canham R. P., Birdsall D. A., Cameron D. G., 1970. Disturbed segregation at the transferring locus of the deer mouse. Genet. Res. 16: 355–377. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

