Genome scanning for conditionally essential genes in Salmonella enterica Serotype Typhimurium
- PMID: 22367088
- PMCID: PMC3346488
- DOI: 10.1128/AEM.06865-11
Genome scanning for conditionally essential genes in Salmonella enterica Serotype Typhimurium
Abstract
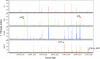
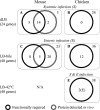
As more whole-genome sequences become available, there is an increasing demand for high-throughput methods that link genes to phenotypes, facilitating discovery of new gene functions. In this study, we describe a new version of the Tn-seq method involving a modified EZ:Tn5 transposon for genome-wide and quantitative mapping of all insertions in a complex mutant library utilizing massively parallel Illumina sequencing. This Tn-seq method was applied to a genome-saturating Salmonella enterica serotype Typhimurium mutant library recovered from selection under 3 different in vitro growth conditions (diluted Luria-Bertani [LB] medium, LB medium plus bile acid, and LB medium at 42°C), mimicking some aspects of host stressors. We identified an overlapping set of 105 protein-coding genes in S. Typhimurium that are conditionally essential under at least one of the above selective conditions. Competition assays using 4 deletion mutants (pyrD, glnL, recD, and STM14_5307) confirmed the phenotypes predicted by Tn-seq data, validating the utility of this approach in discovering new gene functions. With continuously increasing sequencing capacity of next generation sequencing technologies, this robust Tn-seq method will aid in revealing unexplored genetic determinants and the underlying mechanisms of various biological processes in Salmonella and the other approximately 70 bacterial species for which EZ:Tn5 mutagenesis has been established.
Figures




Similar articles
-
Genetic Determinants in Salmonella enterica Serotype Typhimurium Required for Overcoming In Vitro Stressors in the Mimicking Host Environment.Microbiol Spectr. 2021 Dec 22;9(3):e0015521. doi: 10.1128/Spectrum.00155-21. Epub 2021 Dec 8. Microbiol Spectr. 2021. PMID: 34878334 Free PMC article.
-
Preparation of Transposon Library and Tn-Seq Amplicon Library for Salmonella Typhimurium.Methods Mol Biol. 2019;2016:3-15. doi: 10.1007/978-1-4939-9570-7_1. Methods Mol Biol. 2019. PMID: 31197704
-
Genome-Wide Mutagenesis in Borrelia burgdorferi.Methods Mol Biol. 2018;1690:201-223. doi: 10.1007/978-1-4939-7383-5_16. Methods Mol Biol. 2018. PMID: 29032547
-
The bare necessities: Uncovering essential and condition-critical genes with transposon sequencing.Mol Oral Microbiol. 2019 Apr;34(2):39-50. doi: 10.1111/omi.12256. Epub 2019 Mar 1. Mol Oral Microbiol. 2019. PMID: 30739386 Review.
-
Transposon insertion sequencing: a new tool for systems-level analysis of microorganisms.Nat Rev Microbiol. 2013 Jul;11(7):435-42. doi: 10.1038/nrmicro3033. Epub 2013 May 28. Nat Rev Microbiol. 2013. PMID: 23712350 Free PMC article. Review.
Cited by
-
Identifying essential genes in Schaalia odontolytica using a highly-saturated transposon library.bioRxiv [Preprint]. 2024 Jul 18:2024.07.17.604004. doi: 10.1101/2024.07.17.604004. bioRxiv. 2024. Update in: J Bacteriol. 2025 Aug 21;207(8):e0016425. doi: 10.1128/jb.00164-25. PMID: 39071323 Free PMC article. Updated. Preprint.
-
Genome-wide mutant fitness profiling identifies nutritional requirements for optimal growth of Yersinia pestis in deep tissue.mBio. 2014 Aug 19;5(4):e01385-14. doi: 10.1128/mBio.01385-14. mBio. 2014. PMID: 25139902 Free PMC article.
-
Pyrimidine biosynthesis in pathogens - Structures and analysis of dihydroorotases from Yersinia pestis and Vibrio cholerae.Int J Biol Macromol. 2019 Sep 1;136:1176-1187. doi: 10.1016/j.ijbiomac.2019.05.149. Epub 2019 Jun 15. Int J Biol Macromol. 2019. PMID: 31207330 Free PMC article.
-
Generating Transposon Insertion Libraries in Gram-Negative Bacteria for High-Throughput Sequencing.J Vis Exp. 2020 Jul 7;(161):10.3791/61612. doi: 10.3791/61612. J Vis Exp. 2020. PMID: 32716393 Free PMC article.
-
DEG 10, an update of the database of essential genes that includes both protein-coding genes and noncoding genomic elements.Nucleic Acids Res. 2014 Jan;42(Database issue):D574-80. doi: 10.1093/nar/gkt1131. Epub 2013 Nov 15. Nucleic Acids Res. 2014. PMID: 24243843 Free PMC article.
References
-
- Becker D, et al. 2006. Robust Salmonella metabolism limits possibilities for new antimicrobials. Nature 440:303–307 - PubMed
-
- Bossé JT, Zhou L, Kroll JS, Langford PR. 2006. High-throughput identification of conditionally essential genes in bacteria: from STM to TSM. Infect. Disord. Drug Targets 6:241–262 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

