Characterization of DeltaN-Zfp36l2 mutant associated with arrest of early embryonic development and female infertility
- PMID: 22367205
- PMCID: PMC3339969
- DOI: 10.1074/jbc.M111.330837
Characterization of DeltaN-Zfp36l2 mutant associated with arrest of early embryonic development and female infertility
Abstract
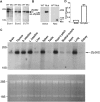
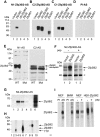
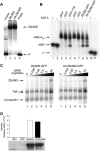
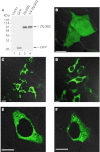
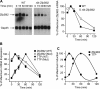
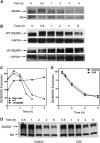
The zinc finger protein 36-like 2, Zfp36l2, has been implicated in female mouse infertility, because an amino-terminal truncation mutation (ΔN-Zfp36l2) leads to two-cell stage arrest of embryos derived from the homozygous mutant female gamete. Zfp36l2 is a member of the tristetraprolin (TTP) family of CCCH tandem zinc finger proteins that can bind to transcripts containing AU-rich elements (ARE), resulting in deadenylation and destabilization of these transcripts. I show here that the mouse Zfp36l2 is composed of two exons and a single intron, encoding a polypeptide of 484 amino acids. I observed that ΔN-Zfp36l2 protein is similar to both wild-type Zfp36l2 and TTP (Zfp36) in that it shuttles between the cytoplasm and nucleus, binds to RNAs containing AREs, and promotes deadenylation of a model ARE transcript in a cell-based co-transfection assay. Surprisingly, in contrast to TTP, Zfp36l2 mRNA and protein were rapidly down-regulated upon LPS exposure in bone marrow-derived macrophages. The ΔN-Zfp36l2 protein was substantially more resistant to stimulus-induced down-regulation than the WT. I postulate that the embryonic arrest linked to the ΔN-Zfp36l2 truncation might be related to its resistance to stimulus-induced down-regulation.
Figures


References
-
- Nie X. F., Maclean K. N., Kumar V., McKay I. A., Bustin S. A. (1995) ERF-2, the human homologue of the murine Tis11d early response gene. Gene 152, 285–286 - PubMed
-
- Maclean K. N., McKay I. A., Bustin S. A. (1998) Differential effects of sodium butyrate on the transcription of the human TIS11 family of early response genes in colorectal cancer cells. Br. J. Biomed. Sci. 55, 184–191 - PubMed
-
- Ramos S. B., Stumpo D. J., Kennington E. A., Phillips R. S., Bock C. B., Ribeiro-Neto F., Blackshear P. J. (2004) The CCCH tandem zinc-finger protein Zfp36l2 is crucial for female fertility and early embryonic development. Development 131, 4883–4893 - PubMed
-
- Blackshear P. J. (2002) Tristetraprolin and other CCCH tandem zinc finger proteins in the regulation of mRNA turnover. Biochem. Soc. Trans. 30, 945–952 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

