Evolutionary genetics of the human Rh blood group system
- PMID: 22367406
- PMCID: PMC3378649
- DOI: 10.1007/s00439-012-1147-5
Evolutionary genetics of the human Rh blood group system
Abstract
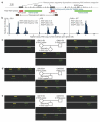
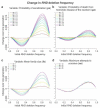
The evolutionary history of variation in the human Rh blood group system, determined by variants in the RHD and RHCE genes, has long been an unresolved puzzle in human genetics. Prior to medical treatments and interventions developed in the last century, the D-positive (RhD positive) children of D-negative (RhD negative) women were at risk for hemolytic disease of the newborn, if the mother produced anti-D antibodies following sensitization to the blood of a previous D-positive child. Given the deleterious fitness consequences of this disease, the appreciable frequencies in European populations of the responsible RHD gene deletion variant (for example, 0.43 in our study) seem surprising. In this study, we used new molecular and genomic data generated from four HapMap population samples to test the idea that positive selection for an as-of-yet unknown fitness benefit of the RHD deletion may have offset the otherwise negative fitness effects of hemolytic disease of the newborn. We found no evidence that positive natural selection affected the frequency of the RHD deletion. Thus, the initial rise to intermediate frequency of the RHD deletion in European populations may simply be explained by genetic drift/founder effect, or by an older or more complex sweep that we are insufficiently powered to detect. However, our simulations recapitulate previous findings that selection on the RHD deletion is frequency dependent and weak or absent near 0.5. Therefore, once such a frequency was achieved, it could have been maintained by a relatively small amount of genetic drift. We unexpectedly observed evidence for positive selection on the C allele of RHCE in non-African populations (on chromosomes with intact copies of the RHD gene) in the form of an unusually high F( ST ) value and the high frequency of a single haplotype carrying the C allele. RhCE function is not well understood, but the C/c antigenic variant is clinically relevant and can result in hemolytic disease of the newborn, albeit much less commonly and severely than that related to the D-negative blood type. Therefore, the potential fitness benefits of the RHCE C allele are currently unknown but merit further exploration.
Figures
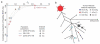

References
-
- Allison AC. The distribution of the sickle-cell trait in East Africa and elsewhere, and its apparent relationship to the incidence of subtertian malaria. Trans R Soc Trop Med Hyg. 1954;48:312–8. - PubMed
-
- Avent ND, Reid ME. The Rh blood group system: a review. Blood. 2000;95:375–87. - PubMed
-
- Barreiro LB, Laval G, Quach H, Patin E, Quintana-Murci L. Natural selection has driven population differentiation in modern humans. Nat Genet. 2008;40:340–5. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

